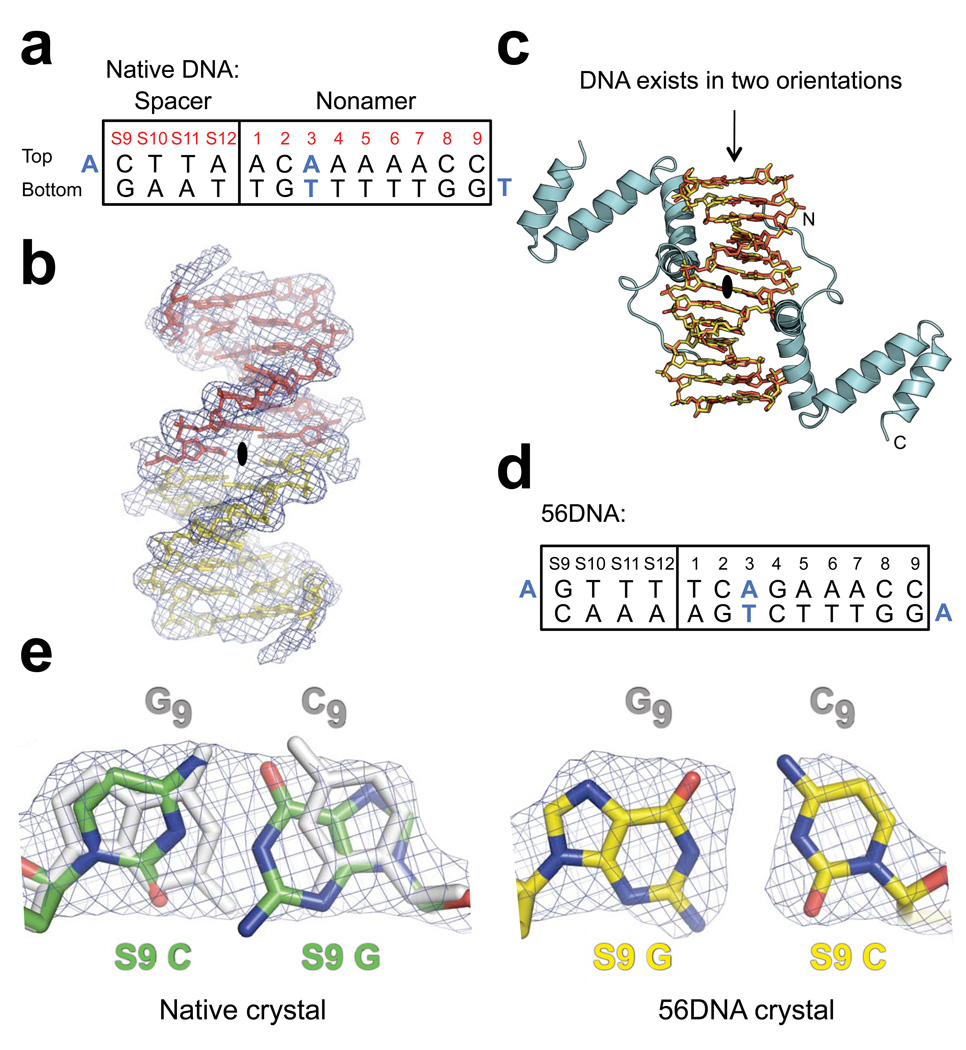
Figure 2.
Electron density maps of DNA in the native and 56DNA crystals. (a) Sequence of the DNA used in native crystals is shown with the consensus nonamer (numbered 1–9) and four base pairs from the consensus 12RSS spacer (numbered S9-S12). Bases coinciding with the crystallographic 2-fold symmetry axis are blue. (b) Solvent-flattened experimental electron density map (mesh) derived from heavy-atom phasing of the native crystal shows continuous electron density extending from the base pair at nonamer position 3 across the 2-fold symmetry axis (black oval; the axis is perpendicular to the page). Two halves of a single 14-mer DNA duplex minus the overhang bases are shown as yellow and red stick models. (c) Application of the crystallographic 2-fold symmetry operator generates a model consistent with the DNA existing in two possible orientations (overlap of yellow and red stick models), each at half occupancy, and the NBD (cyan) bound to the two A•T-rich sites in the DNA. The termini of one NBD are indicated with N and C. Crystals containing a DNA with additional symmetrized bases, 56DNA (d), in complex with the NBD were also obtained. (e) 2Fo-Fc omit electron density maps (mesh) of bases at S9, which were generated by performing simulated annealing refinement without the DNA coordinates, are shown for both the native (green) and 56DNA (yellow) crystals. The corresponding symmetry-mate bases at nonamer position 9 (subscript) are colored gray in both models, but are not visible in the case of 56DNA due to perfect overlap with the S9 base pair. Nitrogen and oxygen atoms are indicated with blue and red, respectively.