Figure 4.
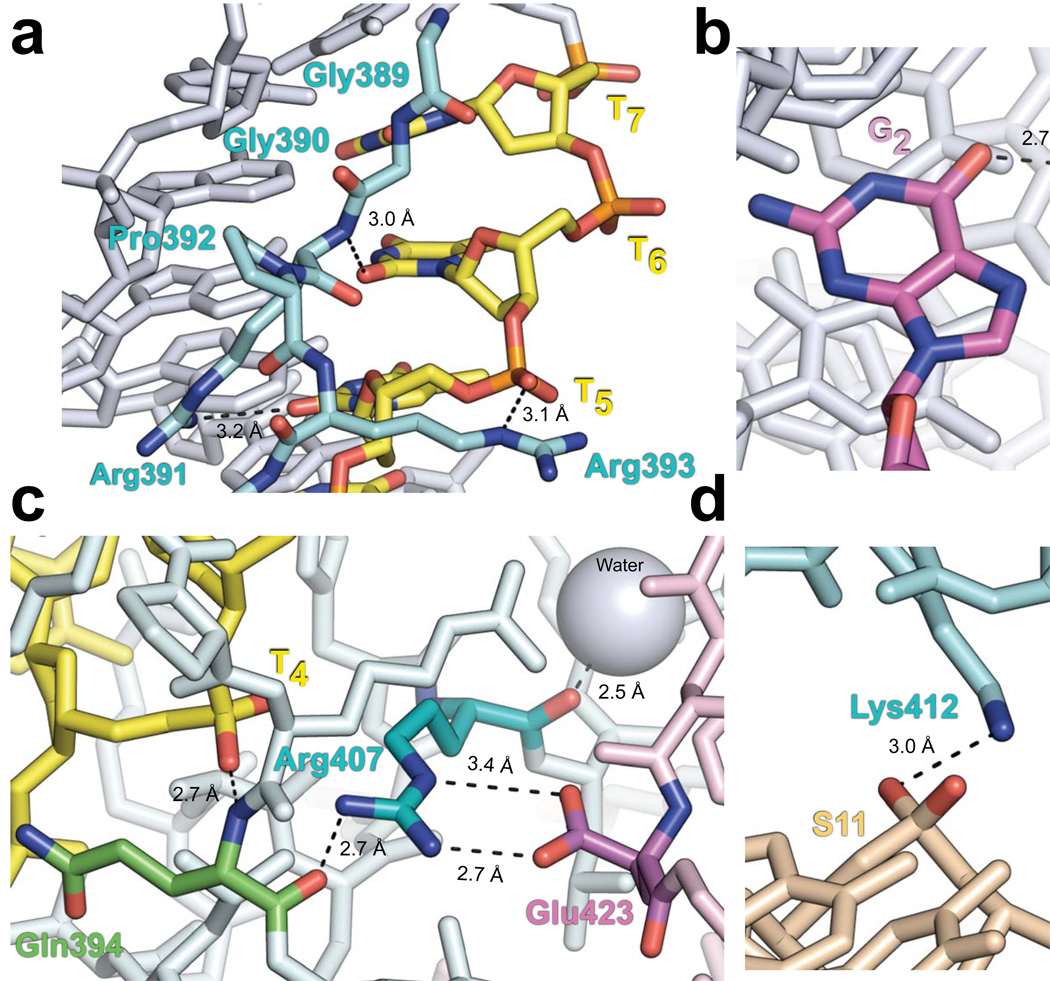
Important protein-DNA and protein-protein contacts in the NBD-DNA crystal structure. Nitrogen, oxygen, and phosphate atoms are indicated with blue, red, and orange, respectively. Dashed lines represent bonds. Subscript denotes the nonamer position. (a) Structure of the GGRPR motif (cyan) in the minor groove of the nonamer is shown with thymines colored yellow, while all other DNA bases are gray. The Arg391 side chain amino group and main chain amide are hydrogen-bonded to the O2 atoms of T5 and T6, respectively. The Arg393 ε-imino group is hydrogen-bonded to the phosphate of T5. (b) Base-specific interaction between the Arg402 side chain (cyan) and the O6 atom of G2 (magenta). (c) The Arg407 guanidino group (cyan) is hydrogen-bonded to the carbonyl of Gln394 (green) in the same subunit (light cyan) and forms a salt bridge with Glu423 (magenta) of the other NBD subunit (light pink). The main chain amide of Gln394 is hydrogen-bonded to the phosphate of T4 (yellow). Additionally, the carbonyl of Arg407 is hydrogen-bonded to a nearby water molecule (gray sphere). (d) In the spacer, the side chains of Lys405 and Lys412 (cyan) form hydrogen bonds with the DNA backbone at positions S10 and S11 (beige), respectively. See also Supplementary Fig. 2.