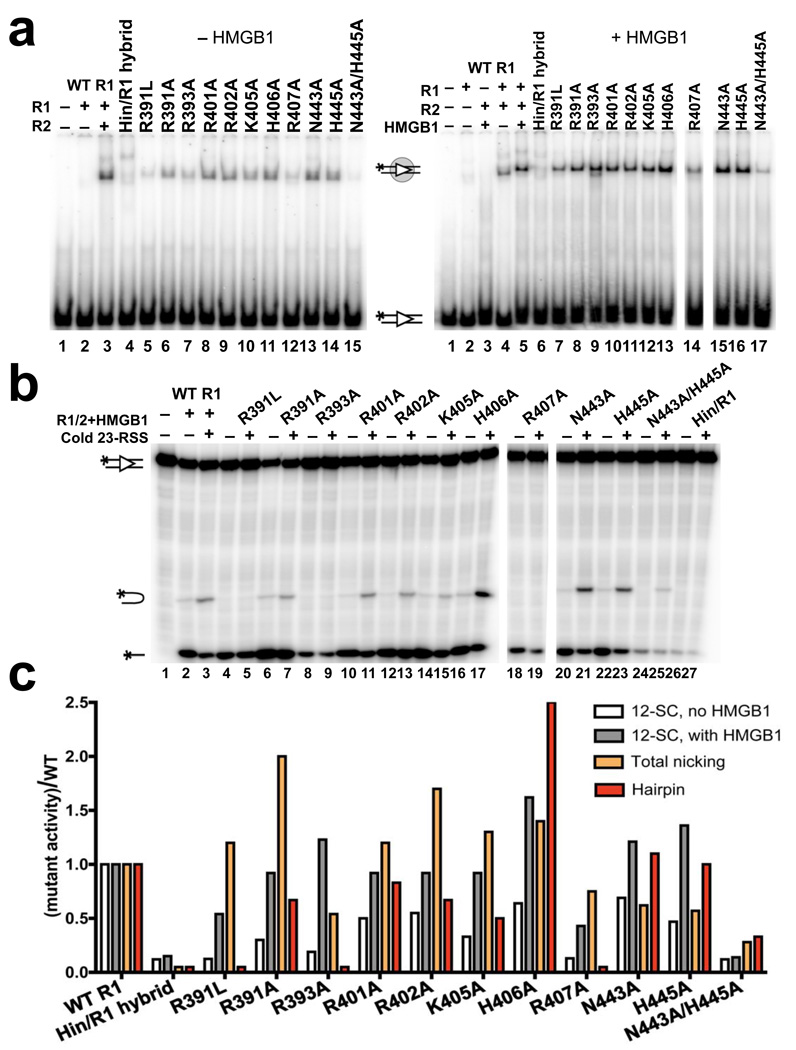
Figure 5.
NBD-DNA contacts are important for DNA binding and hairpin formation. (a) Gel shift analysis of RAG1/2 binding to a 32P-labeled consensus 12RSS substrate in the presence (right gel) or absence (left gel) of HMGB1 in a binding buffer containing 5 mM CaCl2. Proteins were added as indicated above the lanes. All reactions with mutant RAG1 proteins contained RAG2, and the positions of the free and bound substrates are indicated with diagrams. (b) DNA cleavage assay was performed with 32P-labeled consensus 12RSS and a 5-fold molar excess of unlabeled consensus 23RSS in the presence of 1.5 mM MgCl2. Proteins and 23RSS substrate were added as indicated above the lanes. All reactions with mutant RAG1 proteins contained RAG2 and HMGB1. The positions of the nicked product, hairpin product, and input substrate are indicated with diagrams. (c) Quantitation of binding and cleavage results from panels (a) and (b), with the activity of the mutants normalized to that of wild-type RAG1, whose activity was arbitrarily set to one. Similar results were obtained in repeat experiments (data not shown). See also Supplementary Fig. 3.