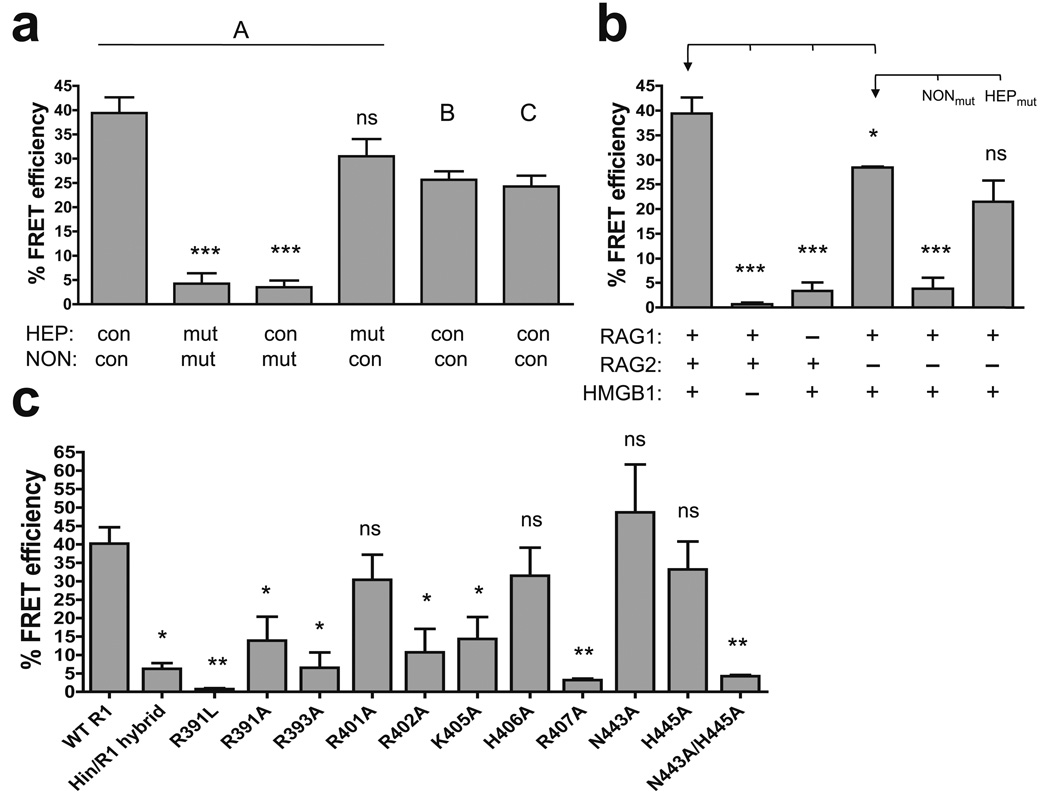
Figure 7.
FRET analysis of the DNA and protein requirements for nonamer synapsis. Average FRET efficiencies of at least two experiments are plotted with error bars indicating the s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (unpaired two-tailed t-test). (a) Energy transfer was assessed with the A, B, and C substrate pairs (as indicated above the bars), and with A substrates in which the heptamer (HEP) and/or nonamer (NON) were mutated in both the 12 and 23 RSSs, as indicated below the bars. mut, mutant; con, consensus. Statistical comparisons were made with the data obtained with the consensus A substrate pair. (b) Energy transfer was assessed using various combinations of RAG1, RAG2, and HMGB1, as indicated below the bars, with the A substrate pair, or the A substrate pair with mutant nonamer (NONmut), or mutant heptamer (HEPmut) sequences. Statistical comparisons (brackets) were made with the data indicated with an arrow. (c) Energy transfer was assessed with the A substrate pair using WT and mutant RAG1 core proteins, as indicated below the bars. Statistical comparisons were made with the data obtained with WT RAG1 core.