Table 4.
Results from a simulation study on 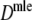 for a range of values of genetic similarity pgs and expected numbers of fragments m1 and m2, 10,000 replicated pairs of AFLP profiles, 1,000 bootstrap resamples, fldFS from A. thaliana with N = 450
for a range of values of genetic similarity pgs and expected numbers of fragments m1 and m2, 10,000 replicated pairs of AFLP profiles, 1,000 bootstrap resamples, fldFS from A. thaliana with N = 450
| Parameter settings | Results for 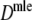
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
 |
Wald ci | Profile likelihood ci | Back transformed Wald ci | |||||||
| pgs | m1 | m2 | Mean | SE | Non-coverage% (too low, too high) |
Length | Non-coverage% (too low, too high) | Length | Non-coverage % (too low, too high) | Length |
| 0.0 | 40 | 40 | 0.0202 | 0.0759 | 0.59 (0.59) | 0.1689 | 1.98 (1.98) | 0.1401 | – | – |
| 70 | 70 | 0.0203 | 0.0651 | 1.18 (1.18) | 0.1477 | 2.35 (2.35) | 0.1275 | – | – | |
| 120 | 120 | 0.0216 | 0.0611 | 1.48 (1.48) | 0.1409 | 2.26 (2.26) | 0.1236 | – | – | |
| 0.1 | 40 | 40 | 0.1004 | 0.0721 | 2.41 (0.97, 1.44) | 0.2320 | 4.49 (2.40, 2.09) | 0.2364 | 5.28 (0, 5.28) | 0.4532 |
| 70 | 70 | 0.1006 | 0.0645 | 3.13 (1.25, 1.88) | 0.2144 | 4.64 (2.29, 2.35) | 0.2164 | 5.51 (0, 5.51) | 0.3974 | |
| 120 | 120 | 0.1001 | 0.0611 | 3.06 (0.94, 2.12) | 0.2056 | 4.94 (2.59, 2.35) | 0.2059 | 5.80 (0, 5.80) | 0.3742 | |
| 0.3 | 40 | 40 | 0.2979 | 0.0807 | 6.27 (3.56, 2.71) | 0.3151 | 5.23 (2.70, 2.53) | 0.3074 | 3.56 (0.02, 3.54) | 0.3123 |
| 70 | 70 | 0.2987 | 0.0690 | 5.77 (2.81, 2.96) | 0.2702 | 5.24 (2.52, 2.72) | 0.2660 | 3.81 (0.15, 3.66) | 0.2670 | |
| 120 | 120 | 0.2985 | 0.0622 | 5.43 (2.69, 2.74) | 0.2436 | 5.08 (2.63, 2.45) | 0.2411 | 3.90 (0.24, 3.66) | 0.2410 | |
| 0.5 | 40 | 40 | 0.4978 | 0.0777 | 5.54 (2.09, 3.45) | 0.3045 | 4.73 (2.42, 2.31) | 0.2948 | 4.09 (1.40, 2.69) | 0.2955 |
| 70 | 70 | 0.4978 | 0.0643 | 5.29 (2.06, 3.23) | 0.2519 | 4.90 (2.36, 2.54) | 0.2495 | 4.38 (1.51, 2.87) | 0.2467 | |
| 120 | 120 | 0.4981 | 0.0560 | 5.08 (2.21, 2.87) | 0.2195 | 4.99 (2.74, 2.25) | 0.2183 | 4.36 (1.76, 2.60) | 0.2161 | |
| 0.7 | 40 | 40 | 0.6974 | 0.0646 | 6.24 (1.66, 4.58) | 0.2532 | 6.39 (3.56, 2.83) | 0.2363 | 4.72 (2.35, 2.37) | 0.2492 |
| 70 | 70 | 0.6989 | 0.0523 | 5.53 (1.75, 3.78) | 0.2049 | 5.01 (2.55, 2.46) | 0.2030 | 4.56 (2.35, 2.21) | 0.2028 | |
| 120 | 120 | 0.6987 | 0.0445 | 5.67 (1.65, 4.02) | 0.1744 | 5.20 (2.43, 2.77) | 0.1741 | 4.88 (2.20, 2.68) | 0.1731 | |
| 0.9* | 40 | 40 | 0.8976 | 0.0382 | 7.74 (0.70, 7.04) | 0.1496 | 12.60 (9.95, 2.65) | 0.1301 | 3.77 (3.04,0.73) | 0.1582 |
| 70 | 70 | 0.8995 | 0.0304 | 6.96 (0.87, 6.09) | 0.1192 | 7.76 (5.14, 2.62) | 0.1092 | 4.24 (2.88, 1.36) | 0.1238 | |
| 120 | 120 | 0.8997 | 0.0255 | 6.83 (1.05, 5.78) | 0.0999 | 5.30 (2.50, 2.80) | 0.0990 | 4.54 (2.87, 1.67) | 0.1026 | |
| 0.95* | 40 | 40 | 0.9491 | 0.0266 | 13.42 (0.22, 13.20) | 0.1007 | 19.34 (15.61, 3.73) | 0.0899 | 6.43 (2.95, 3.48) | 0.1199 |
| * | 70 | 70 | 0.9496 | 0.0208 | 9.96 (0.46, 9.50) | 0.0817 | 12.87 (9.48, 3.39) | 0.0736 | 3.73 (3.20, 0.53) | 0.0923 |
| * | 120 | 120 | 0.9500 | 0.0175 | 9.06 (0.75, 8.31) | 0.0684 | 6.47 (3.42, 3.05) | 0.0659 | 4.10 (3.08, 1.02) | 0.0750 |
| 0.5 | 100 | 50 | 0.4977 | 0.0592 | 5.56 (2.35, 3.21) | 0.2320 | 5.23 (2.66, 2.57) | 0.2301 | 4.71 (1.86, 2.85) | 0.2280 |
| 0.5 | 100 | 80 | 0.4982 | 0.0595 | 5.36 (2.30, 3.06) | 0.2330 | 4.88 (2.57, 2.31) | 0.2314 | 4.54 (1.84, 2.70) | 0.2289 |
| 0.7 | 70 | 40 | 0.6980 | 0.0544 | 5.96 (1.67, 4.29) | 0.2132 | 5.65 (2.77, 2.88) | 0.2054 | 4.97 (2.34, 2.62) | 0.2109 |
| 0.7 | 80 | 70 | 0.6985 | 0.0509 | 5.75 (2.62, 2.21) | 0.1995 | 5.20 (2.84, 2.36) | 0.1983 | 4.83 (2.62, 2.21) | 0.1976 |
Shown are the mean, mean standard error, and properties of three types of confidence intervals: non-coverage percentage (with left and right non-coverage percentages), and mean length of (1) 95% Wald c.i., (2) 95% profile likelihood c.i., and (3) 95% logit-back transformed Wald c.i. At pgs = 0.0, only non-coverage at the right of pgs = 0.0 is considered
* In case pgs = 0.9 (m = 40), or pgs = 0.959 (m = 40, 70, 120) identical pairs of profiles were sampled (10, 348, 53, and 10 times, respectively); in these cases 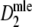 = 1, with standard error 0, and we took logit(pc) = 16 with standard error 0
= 1, with standard error 0, and we took logit(pc) = 16 with standard error 0
