Table 5.
Comparison of bootstrap confidence intervals for pgs from a simulation study on 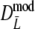 for a range of values of genetic similarity pgs and expected numbers of fragments m1 and m2, 10,000 replicated pairs of AFLP profiles, 1,000 bootstrap resamples, fldFS from A. thaliana with N = 450
for a range of values of genetic similarity pgs and expected numbers of fragments m1 and m2, 10,000 replicated pairs of AFLP profiles, 1,000 bootstrap resamples, fldFS from A. thaliana with N = 450
| Parameter settings | Results for 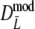 : 95% bootstrap confidence intervals for pgs : 95% bootstrap confidence intervals for pgs
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Percentile bootstrap c.i. | Bias corrected bootstrap c.i. | BCa c.i. | ||||||
| pgs | m1, | m2 | Non-coverage% (too low, too high) | Length (trunc) | Non-coverage% (too low, too high) | Length (trunc) | Non-coverage% (too low, too high) | Length (trunc) |
| 0.0 | 40 | 40 | 6.98 (5.57, 1.41) | 0.2495 (0.1324) | 6.50 (4.73, 1.77) | 0.2517 (0.1386) | 5.34 (3.04, 2.30) | 0.2584 (0.1522) |
| 70 | 70 | 5.68 (3.63, 2.05) | 0.2670 (0.1332) | 5.60 (3.40, 2.20) | 0.2672 (0.1354) | 5.45 (2.88, 2.57) | 0.2680 (0.1396) | |
| 120 | 120 | 5.40 (3.08, 2.32) | 0.2862 (0.1382) | 5.48 (3.23, 2.25) | 0.2862 (0.1381) | 5.38 (3.11, 2.27) | 0.2862 (0.1387) | |
| 0.1 | 40 | 40 | 5.70 (4.16, 1.54) | 0.2893 (0.2369) | 5.21 (3.47, 1.74) | 0.2904 (0.2416) | 4.53 (2.28, 2.25) | 0.2942 (0.2511) |
| 70 | 70 | 5.25 (3.18, 2.07) | 0.2777 (0.2255) | 5.28 (3.01, 2.27) | 0.2778 (0.2778) | 4.93 (2.53, 2.40) | 0.2781 (0.2297) | |
| 120 | 120 | 5.05 (2.95, 2.10) | 0.2796 (0.2220) | 4.98 (2.86, 2.12) | 0.2797 (0.2220) | 4.92 (2.80, 2.12) | 0.2797 (0.2223) | |
| 0.3 | 40 | 40 | 5.49 (3.30, 2.19) | 0.3201 (0.3187) | 5.34 (2.90, 2.44) | 0.3200 (0.3189) | 4.73 (2.16, 2.57) | 0.3205 (0.3197) |
| 70 | 70 | 5.54 (2.89, 2.65) | 0.2781 (0.2776) | 5.51 (2.80, 2.71) | 0.2780 (0.2776) | 5.29 (2.55, 2.74) | 0.2780 (0.2776) | |
| 120 | 120 | 5.19 (2.57, 2.62) | 0.2580 (0.2578) | 5.25 (2.60, 2.65) | 0.2581 (0.2579) | 5.08 (2.58, 2.50) | 0.2582 (0.2579) | |
| 0.5 | 40 | 40 | 4.97 (2.66, 2.31) | 0.3074 | 4.75 (2.44, 2.31) | 0.3072 | 4.30 (2.17, 2.13) | 0.3070 |
| 70 | 70 | 5.09 (2.42, 2.67) | 0.2548 | 5.04 (2.41, 2.63) | 0.2547 | 4.72 (2.30, 2.42) | 0.2548 | |
| 120 | 120 | 5.23 (2.66, 2.57) | 0.2246 | 5.24 (2.62, 2.62) | 0.2246 | 4.99 (2.68, 2.31) | 0.2250 | |
| 0.7 | 40 | 40 | 5.73 (2.41, 3.32) | 0.2568 | 5.48 (2.49, 2.99) | 0.2573 | 4.76 (2.47, 2.29) | 0.2586 |
| 70 | 70 | 5.22 (2.28, 2.94) | 0.2065 | 5.14 (2.32, 2.82) | 0.2068 | 4.76 (2.41, 2.35) | 0.2078 | |
| 120 | 120 | 5.63 (2.32, 3.31) | 0.1760 | 5.53 (2.41, 3.12) | 0.1762 | 5.38 (2.73, 2.65) | 0.1770 | |
| 0.9 | 40 | 40 | 6.30 (1.62, 4.68) | 0.1517 | 5.53 (1.76, 3.77) | 0.1542 | 3.83 (2.18, 1.65) | 0.1613 |
| 70 | 70 | 5.89 (1.58, 4.31) | 0.1202 | 5.33 (1.83, 3.50) | 0.1213 | 4.29 (2.36, 1.93) | 0.1250 | |
| 120 | 120 | 5.64 (1.63, 4.01) | 0.1004 | 5.49 (1.91, 3.58) | 0.1011 | 4.65 (2.36, 2.29) | 0.1032 | |
| 0.95 | 40 | 40 | 9.15 (0.85, 8.30) | 0.1020 | 7.07 (1.04, 6.03) | 0.1060 | 5.16 (1.59, 3.57) | 0.1173 |
| 70 | 70 | 7.63 (1.18, 6.45) | 0.0829 | 6.48 (1.49, 4.99) | 0.0850 | 4.63 (2.22, 2.41) | 0.0907 | |
| 120 | 120 | 6.82 (1.31, 5.51) | 0.0682 | 5.99 (1.48, 4.51) | 0.0708 | 4.37 (2.30, 2.07) | 0.0742 | |
| 0.5 | 100 | 50 | 5.30 (2.79, 2.51) | 0.2337 | 5.37 (2.72, 2.65) | 0.2336 | 5.03 (2.56, 2.47) | 0.2336 |
| 0.5 | 100 | 80 | 5.23 (2.68, 2.55) | 0.2364 | 5.17 (2.65, 2.52) | 0.2365 | 5.03 (2.65, 2.38) | 0.2367 |
| 0.7 | 70 | 40 | 5.62 (2.33, 3.29) | 0.2150 | 5.35 (2.20, 3.15) | 0.2149 | 4.81 (2.17, 2.64) | 0.2157 |
| 0.7 | 80 | 70 | 5.48 (2.54, 2.94) | 0.2009 | 5.51 (2.63, 2.88) | 0.2012 | 5.21 (2.83, 2.38) | 0.2021 |
Shown are non-coverage percentages (with left and right non-coverage percentages) and mean length of (1) 95% percentile bootstrap c.i., (2) 95% bias-corrected bootstrap c.i., and (3) 95% accelerated bias-corrected (BCa) bootstrap c.i
