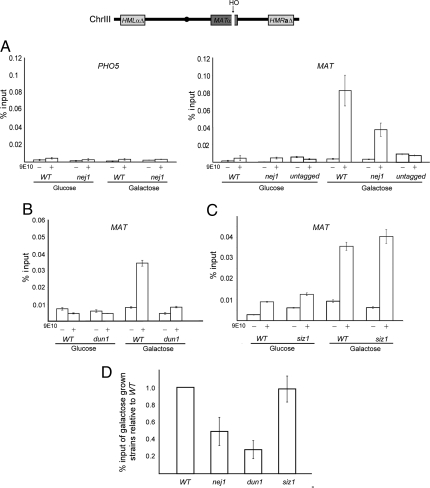
Fig. 2.
Recruitment of Srs2–13XMyc to an induced DNA DSB. Chromatin immunoprecipitation (ChIP) was performed on strain SAY1110 (SRS2-13XMYC) and derivatives using an anti-myc antibody (9E10). Cell extracts were prepared from each strain following growth in glucose or galactose, with galactose resulting in the induction of a single DSB by the HO endonuclease at MAT, schematically shown on top. Following ChIP, associated DNA was quantified by qPCR. Mock immunoprecipitations lacking α-myc (9E10) are included. Shown are representative results of experiments repeated at least 3 times, except the experiment in which 9E10 antiserum was added to an untagged strain (SRS2 JKM115), in which a single experiment was performed. The standard deviations of triplicate qPCR reactions for each strain and condition are included. (A) Amplification of MAT associated DNA approximately 0.2 kb away from the induced DSB, and amplification of nonspecifc DNA immediately upstream of the PHO5 locus of chromosome II in a WT (SAY1110) and nej1 (SAY1126) strains. (B) Identical experiments performed in dun1 (SAY1180) or (C) siz1 (SAY1196) strains. (D) For each of the indicated strains, the amplification of MAT associated DNA following DSB induction from multiple experiments is shown as an average relative to WT. The averages represent 3 independent experiments for the nej1 and dun1 strains, and 2 independent experiments for the siz1 strain. The standard deviation between separate experiments is indicated.