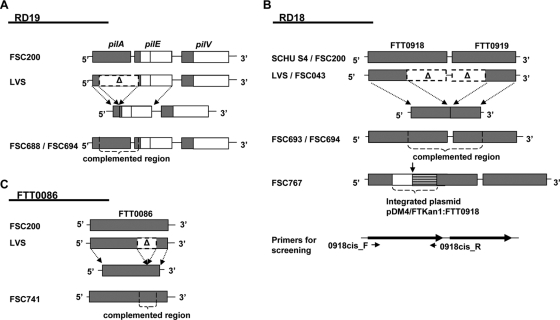
FIG. 1.
Comparison of the RD19, RD18, and FTT0086 regions in different Francisella subspecies and strains. (A) RD19, containing pilA, pilE, and pilV in FSC200 (type B), LVS (type B), FSC688 (LVS2 cis complemented with pilA), and FSC694 (LVS2 cis complemented with pilA and FTT0918). A deletion spanning the pilA gene and the N-terminus-encoding part of pilE results in an in-frame fusion consisting of the N-terminus-encoding part of pilA and the C-terminus-encoding part of pilE in LVS. (B) RD18, containing FTT0918 and FTT0919 in SCHU S4 (type A), FSC200 (type B), LVS (type B), FSC043 (type A), FSC693 (LVS2 cis complemented with FTT0918), FSC694 (LVS2 cis complemented with pilA and FTT0918), and FSC767 (FSC200 with a FTT0918 insertion mutation). In LVS and FSC043, a deletion overlapping the genes FTT0918 and FTT0919 results in an in-frame fusion consisting of the N-terminus-encoding part of FTT0918 and the C-terminus-encoding part of FTT0919. The solid arrow indicates the truncation of the FTT0918 insertion mutant (FSC767). (C) FTT0086 in FSC200 (type B), LVS (type B), and FSC741 (LVS2 cis complemented with FTT0086). In LVS, FTT0086 has undergone a deletion of 93 nucleotides in the middle part of the gene.