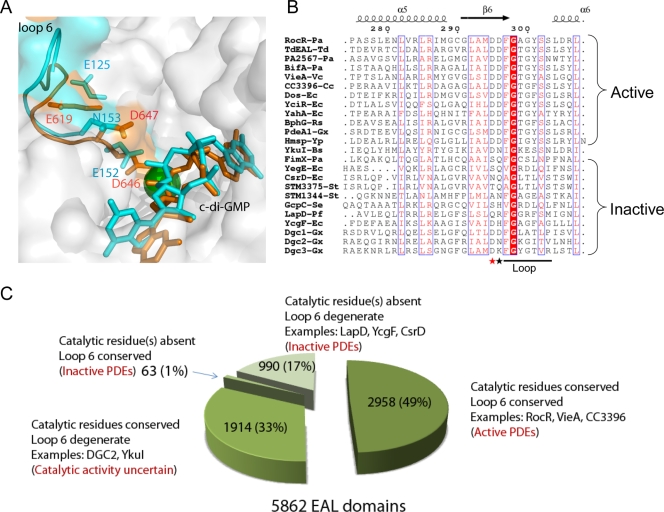
FIG. 7.
Summary of EAL domains. (A) Comparison of loop 6 in active and inactive EAL domains. Loop 6 of TdEAL (Protein Data Bank accession no. 2r6o) and YkuI (Protein Data Bank accession no. 2w27) were highlighted in orange and cyan, respectively. The Mg2+ ion in the TdEAL structure is shown as a green ball, and c-di-GMP is shown as a stick. The corresponding residues of Asp295, Asp296, and Glu268 of RocR are highlighted and labeled (Asp646, Asp 647, and Glu619 for TdEAL; Asp152, Asn153, and Glu125 for YkuI). (B) Comparison of the sequences of the loop among characterized EAL domains. The EAL domains shown are from the following proteins: PA2567, BifA, and FimX from Pseudomonas aeruginosa (indicated by the Pa suffix after the hyphen and protein) (16, 17, 31); TdEAL from Thiobacillus denitrificans (TdEAL-Td); VieA from Vibrio cholerae (VieA-Vc) (38); CC3396 from Crescentus caulobacter (CC3396-Cc) (9); YcgF, YahA, Dos, YciR, CsrD, and YegE from Escherichia coli (indicated by the Ec suffix after the hyphen and protein) (11, 32, 36, 40, 41); BphG from Rhodobacter sphaeroides (BphG-Rs) (39); PdeA1, DGC1, DGC2, and DGC3 from Gluconacetobacter xylinus (indicated by the Gx suffix after the hyphen and protein) (7, 37); HmsP from Yersinia pestis (Hmsp-Yp) (2); GcpC from Salmonella enterica (GcpC-Se) (10); STM1344 and STM3375 from Salmonella enterica serotype Typhimurium (indicated by the St suffix after the hyphen and protein)(33); and LapD from Pseudomonas fluorescens Pf0-1 (LapD-Pf) (24). The sequences were aligned using MultAlin (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html), and the figure was generated using ESPript 2.2 (http://espript.ibcp.fr/ESPript/ESPript/). (C) Pie chart summary of the classification of the 5,862 EAL domains from bacterial genomes according to the conservation of catalytic residues and loop 6.