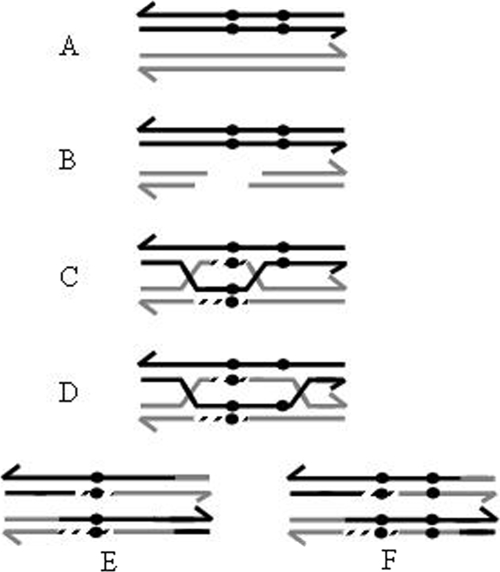
FIG. 1.
Gene conversion under the DSBR model. (A) Two homologous sequences are shown, differing by sequence polymorphisms (black circles). (B) A double-strand break in the recipient homolog is processed (by RecBCD or AddAB) to a gap, generating 3′ tails. (C) After homolog invasion, DNA synthesis (discontinuous lines) fills the gap, and upon ligation, two HJs are formed. At the gap-filling step, gene conversion has occurred because the uncut sequence was the template for gap resynthesis (note the black circles in the cut homolog). (D) HJ migration (by RuvAB, RecG, or RadA) leads to heteroduplex formation. Mismatch repair in the heteroduplex region (mediated by MutS) dictates, depending on the orientation, whether further gene conversion occurs. (E) After HJ resolution, a crossover event with gene conversion to both the gain (black circles in both homologs) and loss of the polymorphisms is generated. (F) Another possible outcome is a crossover event with gene conversion to the gain of both polymorphisms. Only the resolution of HJs leading to crossovers is shown.