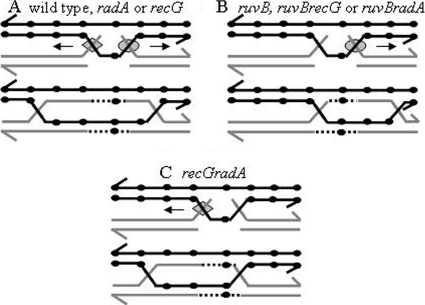
FIG. 5.
Proposal to explain shifting of the most converted sites in HJ mutants. In all panels, two recombining homologs are shown, as well as 3′ ends (half-arrowheads), RFLPs (small black circles), RuvAB (gray diamonds), RecG or RadA (gray circles), and the direction of movement of HJ migration proteins (arrows). (A) HJ migration and gene conversion in the wild type and recG or radA mutant strain. At the top of panel A, 3′ end invasions are processed by RuvAB, while 5′ end invasions are processed by RecG or RadA. These intermediates migrate preferentially in the direction indicated by the small arrows. Given the high processivity of RuvAB, migration toward the 5′ end spans a larger extension than migrations toward the 3′ end. The product of both migration events encompasses a sector located in the middle of the gene. (B) HJ migration and gene conversion in the absence of ruvB (the ruvB, ruvB recG, or ruvB radA mutant strain). Under these circumstances, 3′ end invasions are not processed, leaving only the processing of 5′ invasions by RecG or RadA. Preferential movement of these intermediates toward the 3′ end (top) instigates gene conversion events shifted toward the 3′ end of the gene (bottom). (C) HJ migration and gene conversion in the absence of both recG and radA. In this case, processing of 3′ end invasions by RuvAB provokes the movement of HJs (and hence gene conversion) mostly toward the 5′ end, although migration covering the 3′ end is yet possible, given the high processivity of this complex.