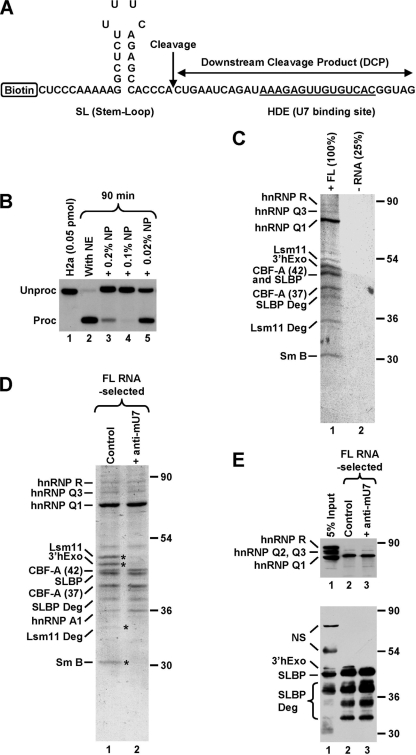
FIG. 1.
Affinity purification of a stable processing complex assembled on histone pre-mRNA. (A) Sequence of the 62-nucleotide FL RNA containing all elements required for 3′-end processing. The cleavage site is indicated with a vertical arrow, and the HDE that base pairs with the U7 snRNA is underlined. The biotin tag is located at the 5′ end of the RNA. (B) In vitro 3′-end processing of a 5′-labeled 86-nucleotide substrate RNA (0.05 pmol/reaction) that is nearly identical with the RNA shown in panel A but lacks the biotin tag and contains additional nucleotides at the 5′ and 3′ ends. Processing was carried out with a mouse nuclear extract (NE) under control conditions (lane 2) or in the presence of the indicated concentrations of NP-40. Lane 1 contains only the input RNA. (C) Coomassie-stained SDS-polyacrylamide gel containing proteins of a mouse nuclear extract adsorbed on streptavidin beads in the presence of 125 pmol of the biotinylated FL RNA (lane 1) or in its absence (lane 2). Processing complexes were formed in the presence of 0.2% NP-40 and subsequently purified on streptavidin beads. The sample lacking the RNA was four times smaller than the RNA-containing sample. Protein identities determined by MS are shown to the left, and size markers are to the right. In addition to FL proteins, MS identified a number of their degradation products. (D) Coomassie-stained SDS-polyacrylamide gel containing proteins of a mouse nuclear extract that associated with the biotinylated FL RNA (125 pmol) in the absence (lane 1) or presence (lane 2) of the anti-mU7 oligonucleotide. Protein bands present only in lane 1 are indicated with asterisks. (E) A fraction of material shown in D was analyzed by Western blotting with anti-hnRNP Q/R (top) or mixed antibodies to 3′hExo and SLBP (bottom). “NS” indicates cross-reacting proteins.