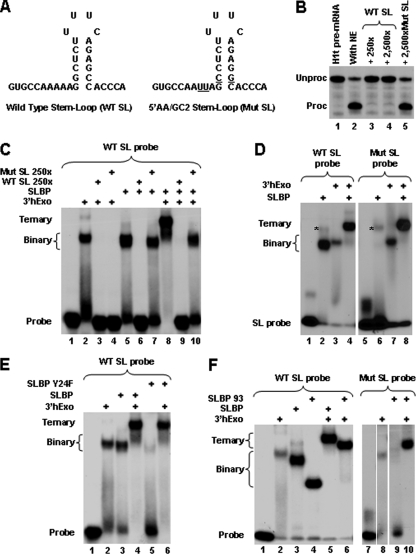
FIG. 3.
Cooperative binding of 3′hExo and SLBP to the SL RNA. (A) Sequences of the WT SL RNA and its mutant (Mut SL), containing nucleotide substitutions (underlined). (B) Ability of a molar excess of the WT and Mut SL RNA competitors to affect cleavage of the H1t histone pre-mRNA by sequestering SLBP present in the nuclear extract. Each competitor was mixed with the 5′-labeled H1t substrate and incubated with a mouse nuclear extract for 1 h. The input H1t pre-mRNA is shown in lane 1. (C) Ability of a molar excess of the WT and Mut SL RNA competitors to affect formation of binary and ternary complexes formed on the 5′-labeled WT SL probe by SLBP and/or 3′hExo. Each competitor was mixed with the 5′-labeled WT SL probe in the presence of 20 mM EDTA and incubated on ice for 30 min with baculovirus-expressed SLBP and/or 3′hExo. Complexes were analyzed with a 6% native polyacrylamide gel. (D) Ability of the 5′-end-labeled Mut SL RNA to form binary and ternary complexes with SLBP and/or 3′hExo. The Mut SL RNA probe was incubated on ice for 30 min with the baculovirus-expressed SLBP and/or 3′hExo in the presence of 20 mM EDTA, and complexes were analyzed with a 6% native polyacrylamide gel. A nonspecific complex formed by a contaminating protein present in the SLBP preparation (lanes 2 and 6) is indicated with an asterisk. (E) Ability of 3′hExo to recruit the Y24F SLBP mutant to the WT SL RNA, giving rise to a ternary complex containing all three components (lane 6). Formation of a binary and ternary complex containing the WT SLPB is shown for comparison in lanes 3 and 4. (F) Mapping of the region in SLBP required for the cooperative interaction with 3′hExo. The ability of the 93-amino-acid region of SLBP to form a ternary complex with 3′hExo and the Mut SL probe is shown (lane 10). Formation of a binary and ternary complex containing the WT SLPB and the 93-amino-acid SLBP mutant with the WT SL probe is shown in lanes 2 to 6.