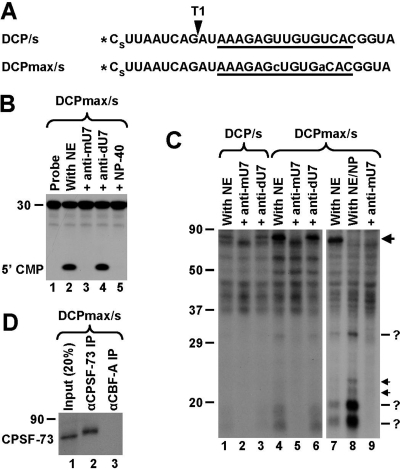
FIG. 4.
U7-dependent interactions in the region between the cleavage site and the HDE. (A) Sequence of the DCP/s and DCPmax/s substrates containing a single radioactive phosphate at the 5′ end (asterisk) and a phosphorothioate modification (s) between nucleotides 1 and 2. The HDE is underlined, and the site recognized by RNase T1 is indicated with an arrowhead. Nucleotide substitutions within the HDE of the DCPmax/s RNA that improve the base pair interaction with the U7 snRNA are indicated with lowercase letters. (B) In vitro degradation of the DCPmax/s RNA. Samples were incubated for 90 min at 32°C either under control conditions (lane 2) or in the presence of the indicated reagents (lanes 3 to 5). The length of the RNA substrate and the position of labeled mononucleotide are indicated to the left. (C) UV-cross-linking using the DCP/s (lanes 1 to 3) and DCPmax/s (lanes 4 to 9) RNAs. Samples were preincubated for 10 min at 32°C either under control conditions or in the presence of the indicated oligonucleotides. Samples in lanes 8 and 9 additionally contained 0.1% NP-40. Following incubation, samples were UV irradiated, treated with RNase T1, and separated in an SDS-polyacrylamide gel. The 80-kDa cross-link at the top of the gel is indicated with the large arrow. Low-molecular-weight U7-dependent cross-links are indicated with question marks (more-intense bands) and arrows (less-intense bands). (D) Immunoprecipitation (IP) of the 80-kDa cross-linked protein with anti-CPSF-73 antibody. A processing reaction mixture containing the 5′-labeled DCPmax/s substrate was UV-irradiated, treated with RNase T1, denatured by boiling in the presence of SDS, and incubated with the anti-CPSF-73 antibody (lane 2) or the anti-CBF-A antibody (lane 3). Precipitated proteins were resolved in an SDS-polyacrylamide gel and detected by autoradiography. The input (20%) is shown in lane 1.