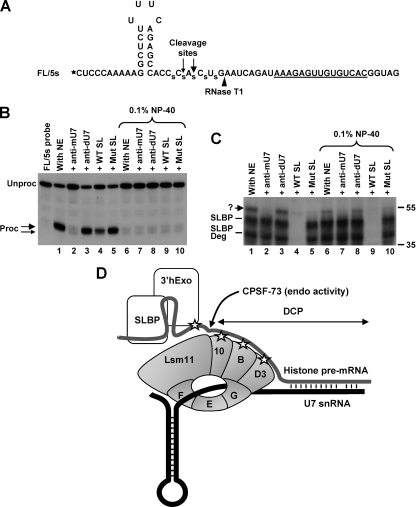
FIG. 6.
U7-dependent interaction in the SL region. (A) Sequence of the FL/5s substrate. The 32P radioactive label at the 5′ end is represented by an asterisk, and the major and minor cleavage sites are indicated with large and small arrows, respectively. The site recognized by RNase T1 is indicated with an arrowhead, and phosphorothioate bonds are represented by “s.” (B) In vitro 3′-end processing of the FL/5s substrate labeled at the 5′ end. Samples were incubated for 90 min at 32°C under control conditions (lanes 1 and 6) or in the presence of a molar excess of the indicated competitors (lanes 2 to 5 and 7 to 10). Samples in lanes 6 to 10 additionally contained 0.1% NP-40. Note that the first lane containing the probe alone is not numbered, so the remaining lanes have the same numbers as the corresponding lanes in panel C. Products generated by cleavage at the major and minor cleavage sites are indicated with large and small arrows, respectively. (C) UV-cross-linking with the FL/5s substrate labeled at the 5′ end. A fraction of each sample shown in panel B, following 10 min of incubation at 32°C, was UV irradiated, treated with RNase T1, and separated in an SDS-polyacrylamide gel. The unknown cross-link is indicated with a question mark. (D) Hypothetical model depicting possible interactions in the stable processing complex assembled on histone pre-mRNA before cleavage. The top (gray) and bottom (black) lines represent histone pre-mRNA and U7 snRNA, respectively. Interactions detected by UV cross-linking are represented by asterisks. The site of cleavage by CPSF-73 is indicated. 3′hExo is actively recruited to the SL structure by the U7 snRNP and possibly by SLBP. After cleavage, 3′hExo tightly associates with the mature 3′ end bound by SLBP.