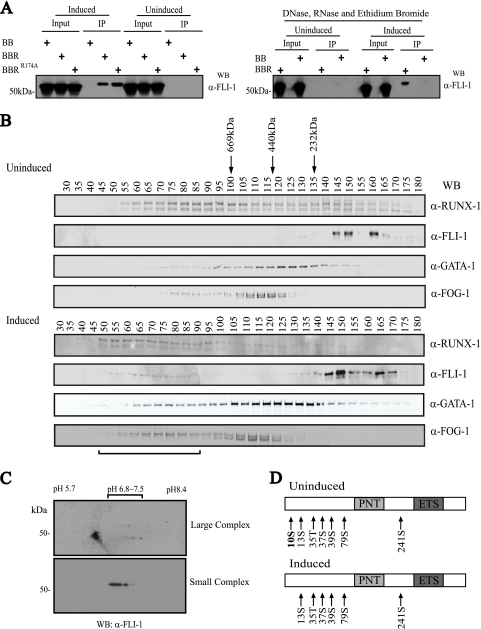
FIG. 5.
Posttranslational modification of FLI-1 correlates with assembly of a large multiprotein complex. (A) Differentiation-dependent and DNA-/RNA-independent interaction between RUNX-1 and FLI-1. (Left) Western blot (WB) for FLI-1 of eluted material, following tandem anti-FLAG-SA affinity purification of FLAG-BioRUNX-1 multiprotein complexes from TPA-induced (3 days) or uninduced L8057 cells. BB, birA and empty FLAG-biotinylation vector; BBR, birA and FLAG-BioRUNX-1; BBRR174A, birA and non-DNA binding mutant FLAG-BioRUNX-1R174A. (Right) The same tandem affinity purification as that for the left panel, but nuclear extracts were treated with DNase, RNase, and ethidium bromide (50 μg/ml) prior to and during co-IP. α-, anti-. (B) Sephacryl 400-gel filtration chromatography of crude nuclear extracts from uninduced (top) and 3-day TPA-induced (bottom) L8057 cells. Western blot analysis of every fifth fraction for RUNX-1, FLI-1, GATA-1, and FOG-1 is shown. Migration positions of globular molecular weight standards are indicated. The void volume corresponds to fractions 19 to 20. Horizontal bar at bottom indicates fractions containing the “large” FLI-1 complex. (C) 2-D gel electrophoresis and anti-FLI-1 Western blot of pooled fractions 66 and 71 (large complex) (top) and pooled fractions 156 and 161 (small complex) (bottom) from the gel filtration chromatography shown in panel B, bottom. (D) Summary of MS phosphorylation analysis of immunopurified FLI-1 from uninduced and induced L8057 cells. Detected phosphorylated residues are indicated. PNT, pointed domain.