Abstract
Murine gammaherpesvirus 68 (MHV-68) replicates robustly in cell culture, providing a model for studying viral genome replication during de novo infection of tumor-associated herpesviruses. We have previously identified a 1.25-kb origin of lytic replication (oriLyt) for MHV-68. To further investigate the molecular mechanism of viral genome replication, we first fine-mapped essential cis-elements from this oriLyt fragment using a transposon-mediated high-density mutagenesis method. The result provided information for us to identify a second oriLyt located towards the left end of MHV-68 genome using a de novo infection-replication assay. We further characterized this left oriLyt by scanning deletion analysis and site-directed mutations, and showed that several CCAAT motifs are essential for oriLyt function, whereas an AT-rich region enhances replication. However, GC-rich repeats are not important cis-element. Moreover, we identified a cellular transcription factor, NF-Y, which binds to CCAAT boxes in EMSA and associates with oriLyt in ChIP assay. Using a dominant negative expression plasmid, we demonstrated that NF-Y plays an important role in mediating MHV-68 genome replication during de novo infection.
Keywords: MHV-68, oriLyt, CCAAT boxes, NF-Y, KSHV
INTRODUCTION
Based on the genome compositions and biological properties, members from herpesvirus family have been classified into three sub-families: α (e.g. herpes simplex virus-1), β (e.g. cytomegalovirus), and γ (e.g. Epstein-Barr virus or EBV). In particular, γ-herpesviruses are involved in tumor development. Two important human pathogens are members of this subfamily. EBV, a γ1-herpesvirus, is associated with lymphomas, nasopharyngeal carcinoma, and other types of malignancies. Kaposi’s sarcoma-associated herpesvirus (KSHV, also known as human herpesvirus 8), a γ2-herpesvirus, is associated with Kaposi’s sarcoma and B cell lymphomas (Stevenson, 2004). Studies on these two human viruses are limited by the lack of efficient de novo infection systems and restricted host range. Murine gammaherpesvirus 68 (MHV-68) also belongs to the γ2 subfamily. In contrast to EBV and KSHV, permissive cell lines are available to study de novo infection as well as latency and reactivation of MHV-68. In addition, MHV-68 can infect laboratory mice and establish productive infection in the lung and latent infection in B lymphocytes, macrophages, dendritic cells, and epithelial cells (Nash et al., 2001; Simas and Efstathiou, 1998). The MHV-68 genome has been completely sequenced, and was found to be closely related to KSHV and EBV (Efstathiou et al., 1990; Virgin et al., 1997). As such, MHV-68 infection of mice has emerged as a model for studying virus-host interactions as well as viral pathogenesis for tumor-associated herpesviruses (Simas and Efstathiou, 1998).
Herpesviruses employ different replication mechanisms to proliferate their genomes during their two modes of infection, lytic replication and latency. During latency, by utilizing cellular DNA replication proteins, EBV initiates its DNA replication at ori-P (a latent replication initiation site) so as to replicate in synchrony with the host genome replication and have the viral genome maintained in host cell in an extrachromosomal manner (Collins et al., 2002; Hu and Renne, 2005). During lytic cycle, the virally encoded DNA replication proteins gather at the origin of lytic replication (oriLyt) to initiate the rolling circle replication process. As a result, “head-to-tail” concatemers are produced (although more complicated “replicative intermediates” have also been reported), followed by being processed at terminal repeats and packaged into progeny virions (Kieff, 2001; Mocarski, 2001; Roizman, 2001).
Previous studies on EBV have revealed that, in addition to cellular proteins, seven viral proteins are also required for viral lytic DNA replication. They are single-stranded DNA binding protein, DNA polymerase, helicase, primase, helicase-associated factor, polymerase processivity factor, and the transcriptional activator ZEBRA (also called Zta or Z protein) serving as an origin-binding protein (OBP) (Fixman, Hayward, and Hayward, 1992; Fixman, Hayward, and Hayward, 1995; Hammerschmidt and Sugden, 1988; Kieff, 2001; Oh, Chittenden, and Levine, 1991; Schepers et al., 2001). In KSHV, functional homologues of six of these proteins have been identified. In addition, the K8 (also called KbZip) protein, which shares similarity with the EBV ZEBRA, has recently been shown to carry out the OBP function (Fixman, Hayward, and Hayward, 1995; Lin et al., 2003; Lin et al., 1999). However, the role of cellular factors in γ-herpesviral DNA replication has been less studied. In MHV-68, genes encoding homologues of six of these proteins have also been identified, but no ZEBRA or K8 homologue has been identified to date, suggesting that unique mechanisms governing MHV-68 lytic DNA replication may exist (Ebrahimi et al., 2003; Martinez-Guzman et al., 2003). Defining cis-sequences required for viral genome replication will thus greatly facilitate studies of MHV-68 DNA replication mechanisms.
In this aspect, two oriLyt regions of KSHV have been identified, which share an almost identical 1.15-kb sequence (>90% sequence identity) and a 600-bp downstream GC-rich repeat sequence in opposite directions (AuCoin et al., 2002; Lin et al., 2003; Wang et al., 2004). We have previously identified an oriLyt region located between ORF66 and ORF73 on MHV-68 genome, and have further mapped the oriLyt to a 1.25-kb region (nt. 100,723–101,974) through deletion mutations (Deng et al., 2004). However, in contrast to KSHV, sequence analysis showed that an extended, almost duplicated copy of the 1.25-kb sequence does not exist in MHV-68. Therefore, if a second copy of oriLyt exists towards the left end of the viral genome, like KSHV, its existence and functionality need to be experimentally tested.
Here we report our identification and functional characterization of a second MHV-68 oriLyt. While this manuscript was in preparation, Adler et al reported identification of a relatively large region which contains a second oriLyt in MHV-68 (Adler et al., 2007). In our independent study, we first employed a high density transposon-mediated mutagenesis method to fine-map cis-elements essential for the MHV-68 right oriLyt. The result provided information for us to identify the left MHV-68 oriLyt using a de novo infection-replication assay. Through systematic deletion analysis and site-directed mutagenesis, several viral cis-elements were found to be essential for viral lytic DNA replication. To our surprise, we found that an ubiquitous cellular transcription factor NF-Y binds to MHV-68 oriLyt sequences and plays an important role in viral genome replication during de novo MHV-68 infection.
RESULTS
High-density mapping of the MHV-68 right oriLyt by transposon-mediated mutagenesis
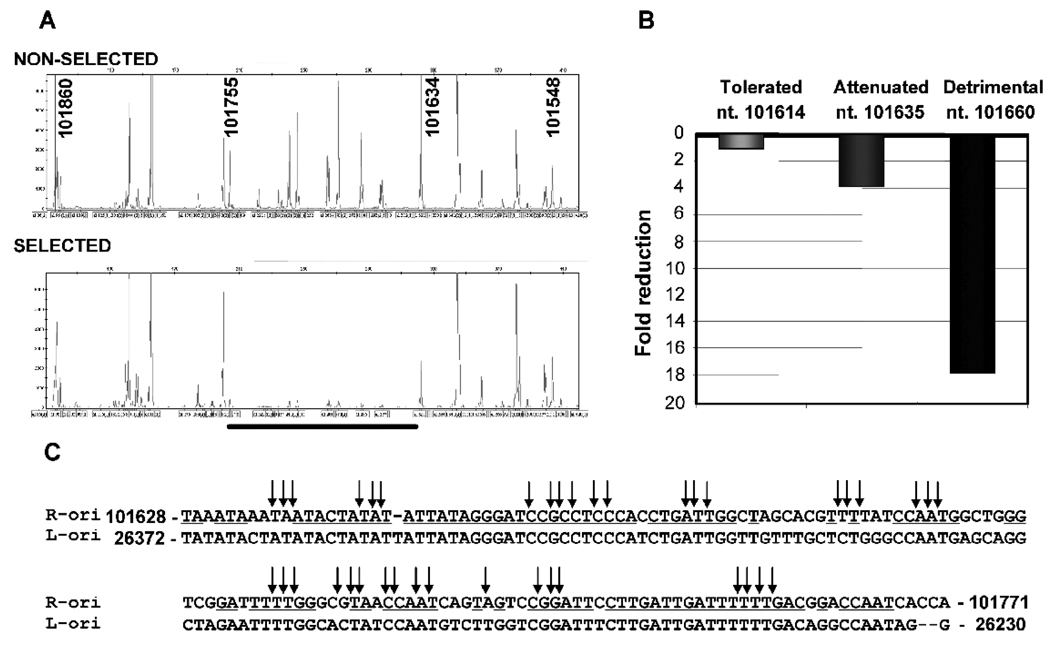
We have previously identified through functional assay a minimal oriLyt of MHV-68 located towards the right end of viral genome (nt. 100,724–101,975). To facilitate understanding of the MHV-68 DNA replication mechanisms, we undertook a high-density transposon-mediated mutagenesis method to map the sequence elements essential for MHV-68 oriLyt replication. The plasmid containing the 1.25-kb right oriLyt was randomly mutagenized using mini-Mu transposons. Subsequent removal of transposon body (1.2-kb in size) from the oriLyt insertion library by restriction digestion resulted in only a 15-bp left in the transposon insertion site. This mutant library was subjected to in vitro selection in 293T cells in the presence of MHV-68 infection. During the selection, the oriLyt plasmids containing 15-bp insertions in the cis-elements that are critical binding sites for viral and cellular trans-factors responsible for oriLyt replication would fail to replicate. The location of 15-bp insertion sites was determined by a genetic foot-printing approach. Briefly, the oriLyt region was enriched by PCR amplification of both the non-selected and the selected oriLyt insertion library. These PCR products were used as a template for a second PCR using a fluorescent-dye labeled insertion-specific primer and one of the oriLyt specific primers. The fluorescent-dye labeled PCR products were run on a capillary sequencer/genotyper. The information regarding the size and the abundance of each insertion specific PCR product were obtained. The genetic foot-printing data were displayed as a chromatogram (Figure 1A) or exported as data file for further data processing and interpretation. For phenotypic assignment, the ratio of each 15-bp insertion peak area was calculated between selected and non-selected oriLyt mutant pools. If a 15-bp insertion resulted in more than 5 fold reduction in replication, this insertion was considered as detrimental, reduction between 2 to 5 fold considered as attenuated phenotype, and less than 2 fold to no change considered as tolerated phenotype (Figure 1B).
Fig. 1.
Identification of essential cis-elements in MHV-68 right oriLyt by a transposon-mediated high-density mutagenesis and genetic foot-printing method. (A) The chromatogram showing the location of 15-bp insertions in the right oriLyt region (nt. 101,860-101,548). Genetic foot-prints of the non-selected (top-panel) and selected (bottom panel) mutant oriLyt pools are shown. Each peak (X-axis) represents the location of 15-bp insertion in the oriLyt region. The fluorescent signal intensity (Y-axis) of each peak indicates the abundance of a particular 15-bp insertion mutant. The region where the 15-bp insertions are detrimental to oriLyt replication is shown (underlined). The MHV-68 genome sequence position is indicated in bold face numbers. Note that the insertion at genome position 101,635 resulted in reduced oriLyt replication. (B) Bar-graph showing the effect of 15-bp insertions on oriLyt replication. The phenotypes of representative insertions are given. (C) The sequence of the right oriLyt region, in which all the 15-bp insertions (arrows) were detrimental for oriLyt replication, is shown in alignment with the sequence of a conserved fragment located in the left oriLyt identified in this study.
Comparison of the genetic foot-printing data obtained from non-selected input pool and in vitro selected pool revealed that insertions between regions corresponding to nt. 101,161 to 101,631 and nt. 101,756 to 101,969 were all tolerated for oriLyt replication. Insertions at genome position nt. 101,634–101,636 resulted in 2–4 fold attenuation of oriLyt replication. All the insertions between nt. 101,643–101,755 were lethal for oriLyt replication (Figure 1A, showing missing peaks in the selected pool), suggesting that the cis-elements located in this region are critical for oriLyt replication. The location of 15-bp insertions in the GC rich repeat region (nt. 100,724–101,160) could not be determined.
Interestingly, although analysis of MHV-68 genome sequences did not reveal a sequence of similar length with extended homology to the 1.25-kb minimal oriLyt we previously mapped (Deng et al., 2004), it did identify a homologous sequence of this short region located towards the left end of genome (Figure 1C), suggesting that this region may be involved in a conserved oriLyt function.
Identification of MHV-68 left oriLyt in a de novo infection-replication assay
Analysis of the right oriLyt by transposon-mediated mutagenesis showed that nt. 101,643–101,755 were essential for oriLyt replication; moreover, a similar sequence was identified between K3 and the 40-bp GC-rich repeats region (Figure 2 A). We thus hypothesized that this region might contain a second oriLyt of MHV-68, since two lytic replication origin sites have been identified in EBV or KSHV (AuCoin et al., 2002; Xue and Griffin, 2007).
Fig. 2.
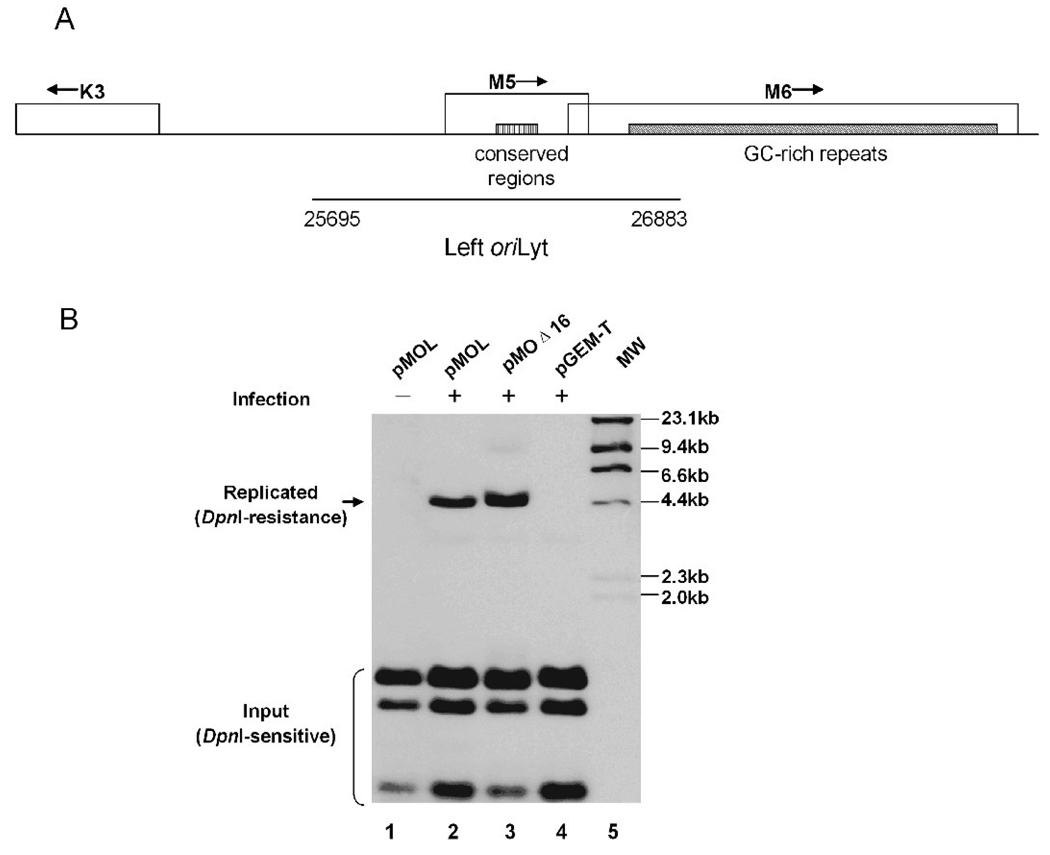
Identification of a second MHV-68 oriLyt through a de novo infection-replication assay. (A) A schematic diagram of the locus on MHV-68 genome which contains the putative left oriLyt. The conserved region between the left and the right oriLyt, as well as viral ORFs in this locus (K3, M5 and M6), are shown. (B) De novo infection-replication assay. The 1.2-kb fragment (nt. 25,695–26,923) was cloned into pGEM-T vector to generate pMOL. pMOL (lanes 1 and 2), pGEM-T (lane 4, negative control), pMOΔ16 (lane 3, positive control) were transfected into 293T cells, followed by infection with wild type MHV-68 at a MOI of 0.05 (lanes 2, 3 and 4). Cellular DNA were extracted and treated with Dpn I and Nde I (for pMOΔ16) or Pst I (for pMOL and pGEM-T). The samples were then run on an agarose gel and analyzed by Southern blotting with DIG labeled pGEM-T as probe. Hind III-digested λ phage DNA served as molecular size markers.
To experimentally test whether this area contained a second functional lytic replication origin for MHV-68, a 1.2-kb DNA sequence (nt. 25,695–26,883) containing the conserved region and flanking fragments was amplified by PCR reaction and cloned into pGEM-T vector to generate pMOL. The resulting plasmid was submitted to a de novo infection-replication assay, as previously described (Deng et al., 2004). Briefly, we transfected the plasmid into 293T cells, and 24 hrs later, infected cells with wild-type MHV-68. Cellular DNA was extracted, digested with Dpn I and a unique cutter, and analyzed by Southern blotting.
As shown in Figure 2B, the vector control, pGEM-T, failed to replicate (lane 4). In contrast, pMOL replicated and yielded a 4.2-kb Dpn I-resistant band (arrow, lane 2), indicating that the 1.2-kb DNA sequence spanning nt. 25,695–26,883 could mediate the replication of plasmid on which it resides. Replication required the presence of viral factors, as pMOL failed to replicate in the absence of viral infection (lane 1). As a positive control, pMOΔ16, containing the minimal right oriLyt of MHV-68 (Deng et al., 2004), replicated as expected (lane 3). Collectively, these data demonstrated that, in addition to the oriLyt that we previously identified, MHV-68 carries a second oriLyt, which is located between nt. 25,695–26,883 towards the left end of the viral genome (termed the left oriLyt).
Systematic deletion analysis of the MHV-68 left oriLyt
A sequence inspection of this 1.2-kb sequence revealed an AT-rich palindrome, several CCAAT boxes, and three copies of the 40-bp GC-rich repeats at the 3’ end. AT-rich region facilitates double-stranded DNA chain melting due to its low annealing temperature, explaining why it is found in most of the viral lytic origins studied to date. The CCAAT box is a widely distributed regulatory sequence present in promoters and enhancers, and the CCAAT related regions can be bound by several cellular transcription regulators, such as CCAAT/enhancing binding protein (C/EBP), CCAAT transcription factor (CTF/NF-1), CCAAT displacement protein (CDP) and NF-Y (CBF, CP1) (Mantovani, 1998). In addition, GC-rich repeats were reported to be an essential region of herpesvirus oriLyt (Pari et al., 2001; Wang et al., 2004).
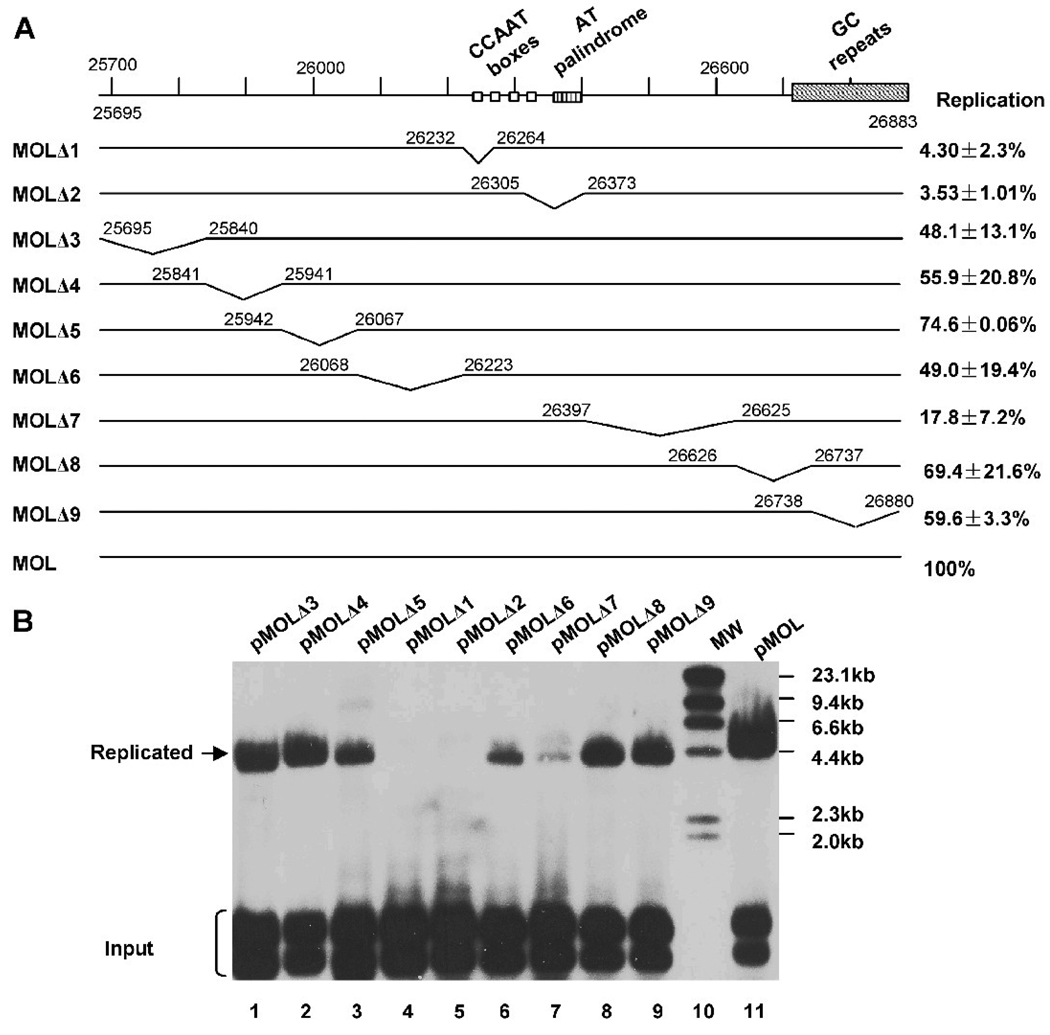
Based on the sequence analysis of the left oriLyt, we took a systematic approach to further map its cis-elements essential for mediating viral DNA replication and to confirm the result of the transposon-mediated mutagenesis of the right oriLyt (Figure 1). A series of deletion mutations were designed and constructed (pMOLΔ1–9, Figure 3A). pMOLΔ1 and pMOLΔ2 were designed to delete the conserved sequences between the left and the right oriLyt, and the other 7 deletion mutations were designed to sequentially delete ~150 bp from the 1.2-kb oriLyt sequence. All constructs were screened by restriction analysis and confirmed by sequencing.
Fig. 3.
Systematic deletion analysis of the MHV-68 left oriLyt. (A) Diagram of deletion plasmids used in this study to analyze the left oriLyt of MHV-68. The replication efficiency of each construct was summarized on the right. Each value represents the average from three independent experiments, with the standard deviation shown. (B) The replication efficiency of these deletion mutations were tested through the de novo infection-replication assay. The replication ability of each plasmid DNA was normalized by taking the ratio of the intensity of the replicated band to that of the input DNA band (specifically, the DpnI-digested band with the slowest mobility), and the replication efficiency of each mutant was calculated by comparing the normalized replication ability to that of pMOL.
We then carried out replication assay to examine the replication efficiency of these deletion plasmids (Figure 3B). The replication ability of each plasmid DNA was normalized with the input DNA, and the replication efficiency of each mutant was calculated by comparing the normalized replication ability to that of pMOL. As shown in Figure 3B, the plasmid replication efficiency retained more than 50% even after deletion of sequences MOL3–6, 8 or 9, suggesting that they are not critical for lytic DNA replication. However, deletion of either of the two conserved sequences (MOL1 and MOL2) abolished DNA replication, indicating that they are essential for oriLyt function. This result also confirmed the data obtained from the high-density mapping. In addition, deletion of sequence MOL7 led to a reduction in replication efficiency to approximately 17.8%, indicating that sequences contained within MOL7 also plays an important role for lytic DNA replication.
Analysis of CCAAT boxes and AT-rich palindrome in MOL1 and MOL2 regions
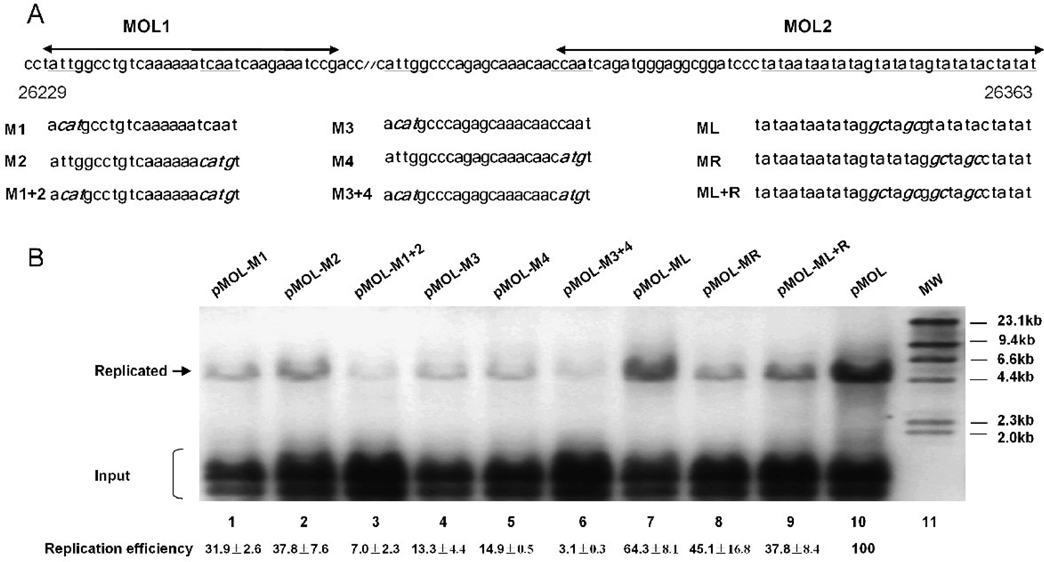
CCAAT boxes have previously been shown to be bound by several cellular transcription regulators, such as C/EBP, NF-1 and NF-Y. To test the importance of each CCAAT box in genome replication of MHV-68, we individually mutated the CCAAT boxes in MOL1 and MOL2 (CCAAT to CATGT, ATTGG to ACATG) (Figure 4A, M1–M4). To further test whether these boxes work in a pair, we also made combined mutations (Figure 4A, M1+2, M3+4). All these mutations were subjected to replication assay to test their replication abilities. Replication efficiency was calculated as described previously. As shown in Figure 4B, mutation of a single CCAAT box markedly reduced oriLyt replication (Figure 4B, lanes 1, 2, 4 and 5), and mutations of two CCAAT boxes abolished the replication (Figure 4B, lanes 3 and 6). These results demonstrated that CCAAT boxes are essential for left oriLyt mediated lytic DNA replication.
Fig. 4.
Analysis of the role of CCAAT boxes and AT-rich palindrome from the left oriLyt. (A) Diagram of the site-directed mutations in MOL1 and MOL2. MOL1 and MOL2 refer to the regions deleted in pMOLΔ1 and pMOLΔ2, respectively. The wide type sequences are shown on the top, and the CCAAT boxes and the AT-rich palindrome are underlined. Sequences containing the mutations (in bold and italic) are shown below. All mutations were confirmed by sequencing. (B) Effect of the site-directed mutations on oriLyt-dependent DNA replication. All mutations were subjected to replication assay and Southern blotting to test their replication abilities. Replication efficiencies shown at the bottom are the average results from three independent experiments.
In order to explore whether one or both arms of the AT-rich palindrome is essential for DNA replication, we also made single and combined mutations of the AT-rich palindrome. Three site-directed mutations were constructed: two contained mutations in either arm and one in both arms (ATATAT to GCATGC) (Figure 4A, right). Results from replication assay showed that mutations of either arm of the AT-rich palindrome affected the replication efficiency slightly (Figure 4B, lanes 7 and 8), whereas mutations of both arms led to a more severe reduction in DNA replication (Figure 4B, lane 9). These results indicated that the AT-rich palindrome plays a role in left oriLyt mediated DNA replication; however it is an auxiliary element rather than an essential one.
GC-rich repeats are not essential for MHV-68 oriLyt function
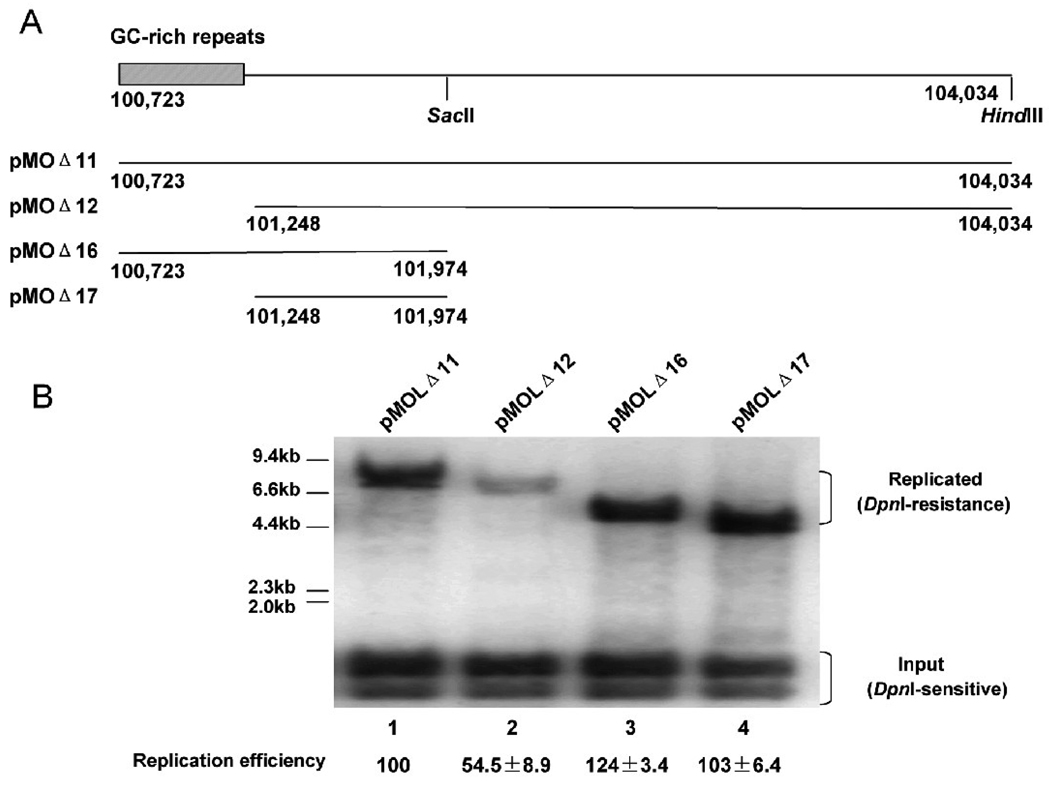
Studies on other herpesvirus oriLyt such as the KSHV oriLyt revealed that GC-rich repeats are essential for lytic DNA replication; excision of the whole GC-rich repeats markedly decreased lytic DNA replication (AuCoin et al., 2002). However, in our study, deletion of the 40-bp GC-rich repeats located at the right end of MHV-68 left oriLyt region did not have much effect on oriLyt function (Figure 3B, pMOLΔ9). It only reduced the replication efficiency to 60%, suggesting that the GC-rich repeats are not essential for lytic DNA replication in MHV-68. Since there are 100-bp GC-rich repeats located adjacent to the left end of MHV-68 right oriLyt, we next examined whether the GC-rich repeats are essential for function of the right oriLyt. In our previous study, we mapped the minimal right oriLyt to a 1.25-kb fragment as contained within pMOΔ16. As shown in Figure 5A, pMOΔ11 and pMOΔ12 have the same 3’ end, whereas pMOΔ16 and pMOΔ11 have the same 5’end. We therefore constructed pMOΔ17 by removing the GC-repeat sequence from pMOΔ12 (Figure 5A). Replication efficiency of pMOΔ17, pMOΔ11, pMOΔ12 and pMOΔ16 were tested through the replication assay (Figure 5B). pMOΔ17 had similar replication efficiency as pMOΔ11 and pMOΔ16, indicating that deletion of the 100-bp GC-rich repeats had little effect on the efficiency of MHV-68 right oriLyt. Taken together with the fact that deletion of the GC-rich repeats in either the left (pMOLΔ9) or the right oriLyt region had no significant effect on oriLyt function or efficiency, we conclude that the GC-rich repeats are non-essential for MHV-68 oriLyt and only serve as an auxiliary element.
Fig. 5.
The 100-bp GC-rich repeats are non-essential for MHV-68 right oriLyt. (A) Diagram of the constructs containing the right oriLyt. Construction of pMOΔ11/12/16 were described previously (Deng et al., 2004). pMOΔ17 was generated by digestion of pMOΔ12 with Sac II and Hind III, followed by self-ligation. pMOΔ12 and pMOΔ17 contain no 100-bp GC-rich repeats. (B) Plasmids were subjected to replication assay and Southern blotting to test their replication abilities.
Cellular factor NF-Y binds to CCAAT boxes in oriLyt in vitro and in vivo
To further investigate the molecular mechanisms governing MHV-68 lytic genome replication, we sought to identify trans-factors involved in MHV-68 genome replication. Since our mutational analysis indicated that CCAAT boxes were essential for oriLyt function, we first focused on identifying cellular factor(s) that plays a role in MHV-68 DNA replication through association with the CCAAT boxes. A number of transcription factors are reported to recognize the CCAAT motif, but subsequent studies have shown that there is a divergence in the recognition sequence of CCAAT, and only NF-Y absolutely requires the CCAAT pentanucleotide (Liberati et al., 1999; Mantovani, 1998). NF-Y has three subunits: A, B, and C. To construct FLAG-tagged NF-Y subunit expression plasmids, a linker containing the FLAG epitope sequence was first inserted into the EcoR I site of vector pSG-5 (Stratagene) to derive pSGFlag. The cDNA sequences of NF-Y subunits were then amplified by PCR, and cloned into the pSGFlag vector to generate pSGFlag-NFYA, pSGFlag-NFYB and pSGFlag-NFYC. Expression of the three plasmids was confirmed by western blotting (data not shown).
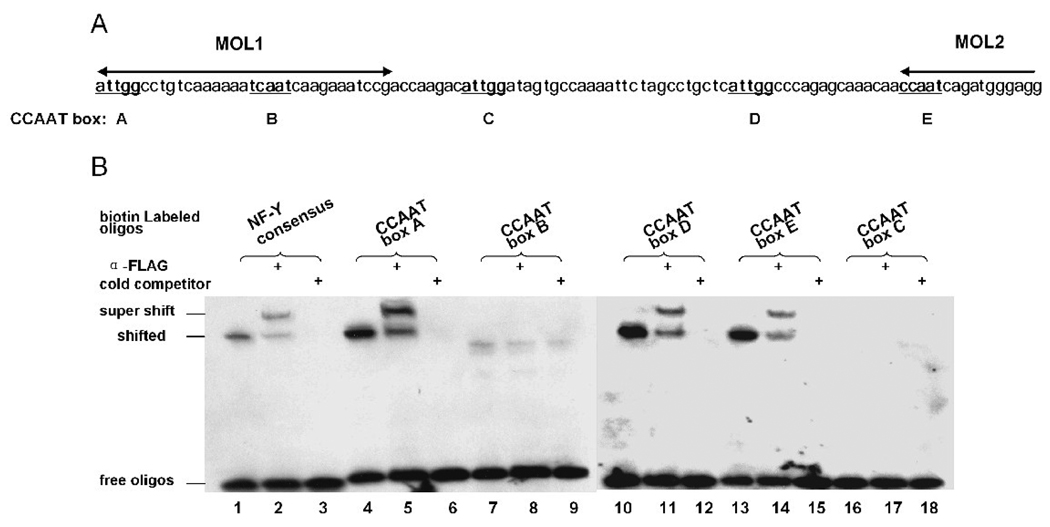
To test whether NF-Y could bind to the CCAAT boxes from the left oriLyt, five biotin end-labeled double-stranded oligonucleotides containing the CCAAT pentanucleotide and 22-nt flanking sequence from the MHV-68 oriLyt region were synthesized (Figure 6A). CCAAT-boxes A and B lie in the region of MOL1, boxes D and E lie in the region of MOL2, and box C lies in the sequence between MOL1 and MOL2. EMSA was performed with nuclear extract prepared from 293T cells previously transfected with pSGFlag-NFYs. NF-Y bound to CCAAT boxes A, D and E as well as the consensus sequence (Figure 6B, lanes 1, 4, 10 and 13), but did not bind to box B or C (Figure 6B, lanes 7 and 16). To confirm the specificity of NF-Y binding, we conducted supershift as well as competition experiments. A monoclonal antibody against the FLAG epitope successfully shifted the bound complex (Figure 6B, lanes 2, 5, 8, 11, 14 and 17). Addition of unlabeled NF-Y consensus oligos to the reaction resulted in disappearance of the bound complexes, again demonstrating specificity of NF-Y binding (Figure 6B, lanes 3, 6, 9, 12, 15, and 18).
Fig. 6.
NF-Y binds to CCAAT boxes in EMSA. (A) Illustration of the CCAAT boxes in MOL1 and MOL2. (B) NF-Y binds to three CCAAT boxes from the left oriLyt. Nuclear extract was harvested from 293T cells co-transfected with pSGFlag-NFYA/B/C, and used in binding reactions with biotin-labeled oligos. In supershift reactions, a monoclonal antibody against FLAG epitope was added; in cold competition reactions, 200-fold unlabeled NF-Y consensus oligos were added. Biotin-labeled NF-Y consensus oligos was used as a positive control for NF-Y binding.
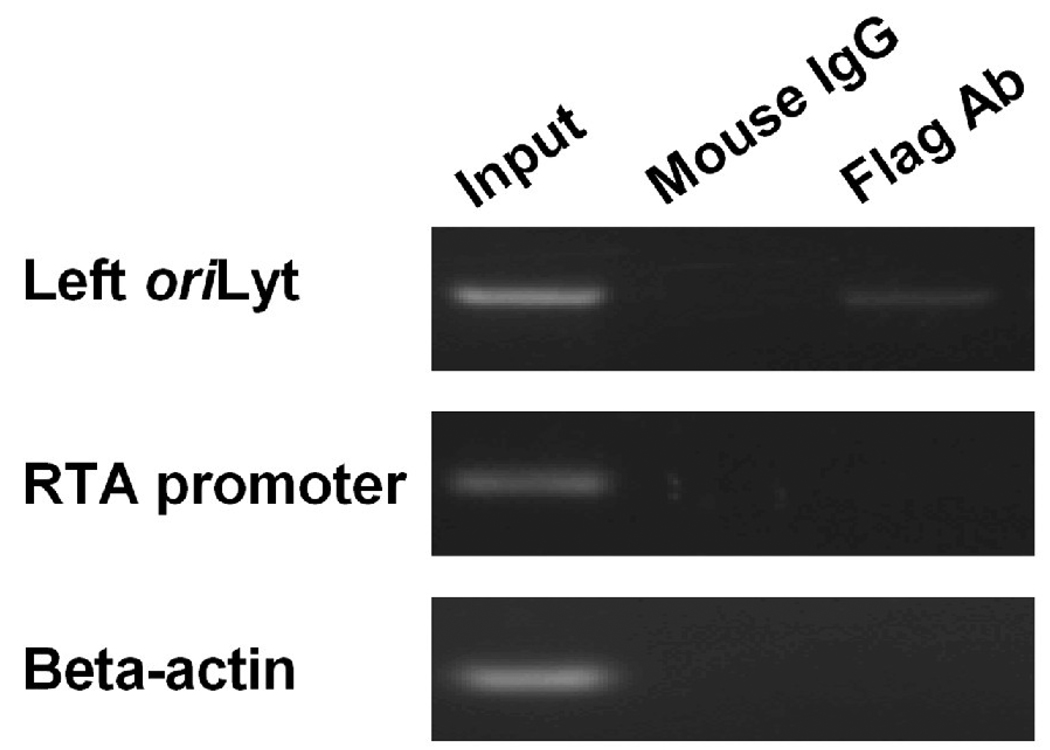
Results of EMSA assay showed that NF-Y could bind to three CCAAT boxes from the left oriLyt in vitro. To examine whether NF-Y could also bind to the CCAAT boxes on viral genome in vivo, chromatin immuno-precipitation (ChIP) assay was employed. Briefly, 293T cells were transfected with FLAG-tagged NF-Y expression plasmids, and 24 hrs later, cells were infected with wild type MHV-68 at a MOI of 5. Twelve hours post-infection, DNA/protein complexes were cross-linked and precipitated with a monoclonal anti-FLAG antibody or an isotype-matched mouse monoclonal antibody. After reversal of cross-linking, the precipitated DNA fragments were PCR amplified with primers specific to the MHV-68 left oriLyt sequences. A specific PCR product was detected when the anti-FLAG antibody was used for pull-down, but not when the control antibody was used (Figure 7, top panel). Moreover, PCR reactions using primers specific for the viral RTA promoter or the cellular beta-actin coding region, which do not contain NF-Y binding sites, yielded no products,(Figure 7, middle and bottom panels). This result demonstrated that NF-Y specifically bound to the left oriLyt region on MHV-68 genome in vivo.
Fig. 7.
NF-Y associates with the left oriLyt on MHV-68 genome in ChIP assay. 293T cells were transfected with pSGFlag-NFYs followed by infection with MHV-68 at a MOI of 5. DNA/protein complexes were cross-linked at 12 hrs post-infection. Sheared DNA/protein complexes were precipitated with a monoclonal antibody against FLAG epitope, and the precipitated DNA was amplified by PCR with primers specific for the left oriLyt, MHV-68 RTA promoter, or beta-actin coding region. Negative controls were carried out with an isotype-matched mouse IgG antibody.
NF-Y plays an important role in the lytic replication of MHV-68
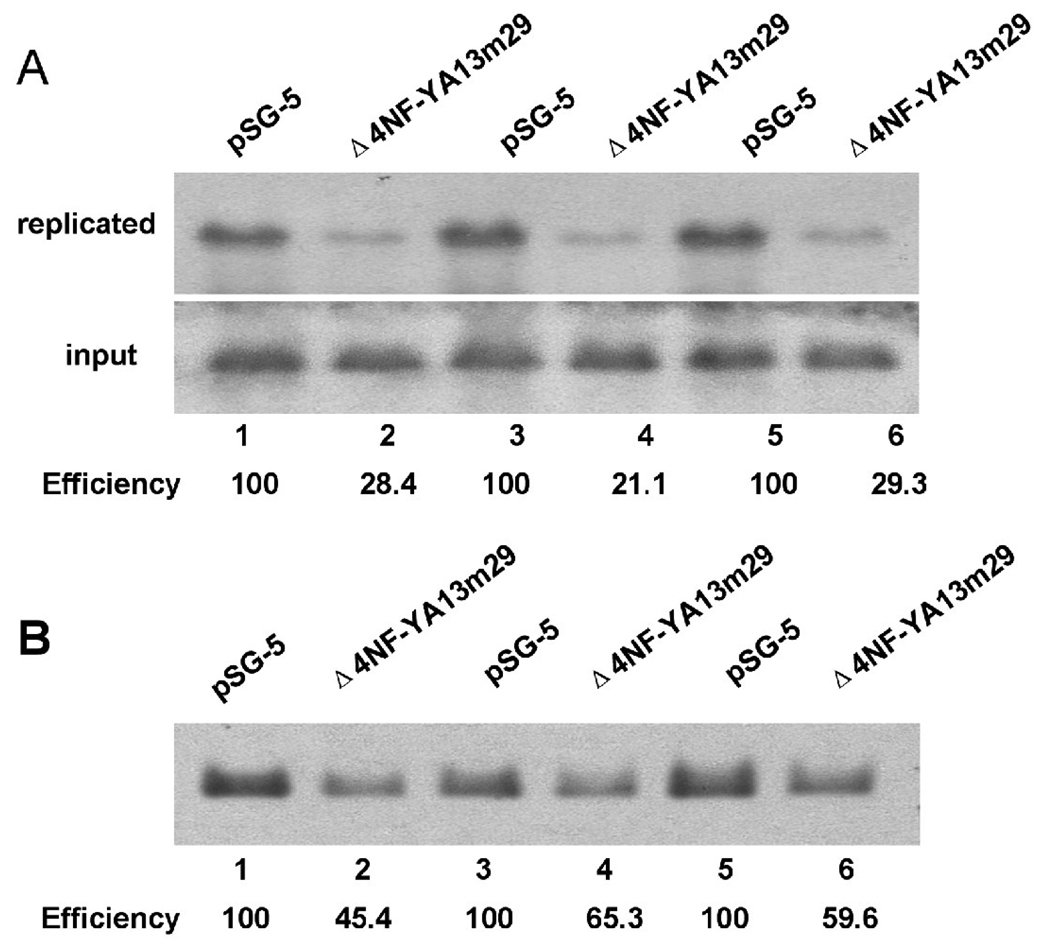
To evaluate the functional significance of NF-Y complex association with the left oriLyt of MHV-68 in vivo, we conducted a knock-down experiment using a dominant negative mutant of NF-Y. This mutated form of NF-YA, expressed from plasmid Δ4NF-YA13m29 (kindly provided by Dr. Mantovani) (Mantovani et al., 1994), sequesters the NF-YB and NF-YC subunits in defective complexes which are unable to bind DNA, and thus functions as a dominant negative repressor of NF-Y/DNA complex formation and of NF-Y dependent functions such as gene regulation (Farina et al., 1999; Mantovani et al., 1994; Wang et al., 1999). 293T cells were co-transfected with pMOL and Δ4NF-YA13m29 (or an empty vector pSG5 as a control), infected with MHV-68, and the replication assay was carried out. Three independent experiments were performed, and a representative result was shown. The replication efficiency of pMOL in the presence of dominant negative NF-Y was significantly lower (Figure 8A, lanes 2, 4 and 6) than that from the control sample (Figure 8A, lanes 1, 3 and 5). To further confirm the role of NF-Y in mediating MHV-68 genome replication during de novo infection, we transfected 293T cells with plasmid Δ4NF-YA13m29 or pSG5, followed by infection with MHV-68. A probe against the viral genome terminal repeat region was used in Southern blotting, to examine the replication efficiency of viral genome. As shown in Figure 8B, expression of the dominant negative form of NF-Y also decreased the replication efficiency of MHV-68 genome (lanes 2, 4 and 6), compared to vector controls (lanes 1, 3 and 5). These results demonstrated that a functional NF-Y complex is required for maximal MHV-68 oriLyt activity.
Fig. 8.
A dominant negative form of NF-Y suppresses the replication of MHV-68 left oriLyt. (A) 293T cells were co-transfected with pMOL and Δ4NF-YA13m29 (or vector pSG-5 as a control) followed by infection with MHV-68 virus at a MOI of 0.05. Total cellular DNA was extracted, digested with Dpn I and a linearizing enzyme to examine newly replicated plasmid DNA (or the linearizing enzyme only, to examine input and newly replicated plasmid DNA), and subjected to Southern blotting. Lanes 1 and 2, 3 and 4, and 5 and 6 represent samples from three independent experiments. (B) 293T cells were transfected with Δ4NF-YA13m29 (or vector pSG-5) followed by infection with MHV-68 at a MOI of 0.05. Total cellular DNA was extracted, digested with Not I, and subjected to Southern blotting with a probe against the terminal repeat region from MHV-68 genome. Lanes 1 and 2, 3 and 4, and 5 and 6 represent samples from three independent experiments.
DISCUSSION
We previously identified the first oriLyt of MHV-68 which is located towards the right end of viral genome. In this study, we fine mapped the right oriLyt of MHV-68 by transposon-mediated mutagenesis, and the results showed that sequences contained within nt. 101,643–101,755 are essential for oriLyt replication. Further analysis of MHV-68 sequences showed that a homologous sequence exists between K3 and the 40bp GC-rich repeats region. A 1.2-kb fragment (nt. 25,695–26,883) containing this homologous sequence indeed conferred oriLyt function in a de novo infection-replication assay. We have thus identified a second oriLyt for MHV-68.
Systematic analysis of the left oriLyt by deletion and site-directed mutations showed that three CCAAT boxes are essential for left oriLyt function, but the AT-rich palindrome only functions as an auxiliary sequence. The 229-bp MOL7 region also contributes to the full replication efficiency of left oriLyt. In addition, deletion of the GC-rich repeats from either the left or the right MHV-68 oriLyt sequence had little effect on the lytic DNA replication efficiency. Moreover, EMSA and ChIP results showed that the cellular factor NF-Y binds to the CCAAT boxes from MHV-68 left oriLyt region in vitro and in vivo. The results of site-directed mutagenesis of CCAAT boxes and the dominant negative assay indicated that NF-Y binding to the essential CCAAT boxes in MOL1 and MOL2 regions play an important role in lytic DNA replication mediated by the left oriLyt.
Comparison of oriLyt from gammaherpesviruses
The lytic DNA replication of EBV and KSHV has been extensively studied. During the viral lytic cycle, multiple rounds of DNA replication are initiated from the oriLyt regions. Two copies of oriLyt have been identified in EBV and KSHV respectively, one towards the left end of viral genome and the other towards the right end. However, these two human herpesviruses both lack efficient experimental de novo infection systems, and their copies of oriLyt were all identified through replication assay supported by reactivation of latent viruses. In contrast, MHV-68 oriLyt could be identified and examined through replication assay driven by de novo infections. Using this de novo infection-replication assay, we have identified and characterized two MHV-68 oriLyt [(Deng et al., 2004), and this study]. Identification of the second oriLyt is also reported by an independent study, which showed that a plasmid containing a 6.9-kb fragment from the MHV-68 genome (nt. 21,383–28,336) was sufficient to mediate oriLyt function in the de novo infection-replication assay (Adler et al., 2007). By making deletion mutants in the context of viral genome, Adler et al have also shown that the fragment spanning nucleotides 26,059 to 28,191 contains cis-elements essential for viral genome replication. Although the region identified in their work is much larger than that in ours (nucleotides 25,695–26,883), their result is consistent with our data from the systematic deletion analysis demonstrating that the fragment spanning nucleotides 26,232 to 26,373 (MOL1 and MOL2) is essential for the function of left oriLyt (Figure 3).
In the case of KSHV, which also belongs to γ2-herpesviruses like MHV-68, the two copies of oriLyt regions share an almost identical 1.15-kb sequence (homology >90%) and a 600-bp downstream GC-rich repeats sequence in opposite directions. Either of the two copies could confer oriLyt function in a plasmid-based assay, however, using KSHV BAC mutants, Xu et al have found that although the presence of oriLyt-L was sufficient to propagate the viral genome, oriLyt-R alone failed to direct the amplification of viral DNA in the context of viral genome (Xu, Rodriguez-Huete, and Pari, 2006). In comparison, the two oriLyt regions on MHV-68 genome do not share as high a similarity as those of KSHV. Instead, they only have one small region of high homology as identified by our high-density transposon-mediated mutagenesis analysis. Aside from the differences between the oriLyt from MHV-68 and KSHV, many similarities also exist. For instance, they all share a similar composition, with a GC-rich repeat region locating at one end, an AT-rich palindrome and several CCAAT boxes in the core regions of oriLyt. As revealed by our study, each motif in MHV-68 oriLyt contributed differently to its function: mutation of the AT-rich palindrome greatly reduced the replication efficiency, whereas mutation of either pair of CCAAT boxes abolished DNA replication; in contrast, deletion of GC-rich repeats had little impact on the function of either the left or the right oriLyt.
Studies on KSHV oriLyt have shown that the AT-rich palindrome is critical for lytic DNA replication - mutation of the 18-bp AT-rich palindrome abolished the oriLyt function (Wang et al., 2004). However, in our study of MHV-68 oriLyt, introducing 8-bp mutations into the AT-rich palindrome only reduced the DNA replication efficiency to half (pMOL-ML+R, Figure 4B, lane 9). One possible explanation for this difference is that the AT-rich palindrome in the left oriLyt of MHV-68 genome is longer than that in KSHV, and mutation of 8-bp was not sufficient to entirely reduce the function of the AT palindrome. In addition, we noted that mutation of both arms of the AT-rich palindrome reduced the replication efficiency to 37.8%, whereas mutation of either arm reduce replication efficiency to 64.3% or 45.1% compared to wild type, indicating additive effect of both arms. This result suggested that this AT-rich sequence may not function by forming a palindrome.
The GC-rich repeats are indispensable to KSHV oriLyt; deletion of GC-rich repeats from the KSHV oriLyt resulted in little lytic DNA replication function. However, our studies of MHV-68 showed that high replication efficiency was still preserved even though the whole GC-rich repeats were deleted from either the left or the right oriLyt. In addition to the more robust nature of the MHV-68 de novo infection-replication assay, it is also possible that the core region of MHV-68 oriLyt has a much higher DNA replication initiation ability compared to that of KSHV oriLyt, so it does not have to rely on the auxiliary function of GC-rich repeats.
Cellular factor NF-Y binds to CCAAT motifs and plays an important role in MHV-68 lytic genome replication
To provide more insights into the molecular mechanisms involved in MHV-68 genome replication, we searched for trans-factors mediating viral DNA replication. Indeed, we showed through EMSA that the cellular transcription factor NF-Y bound to CCAAT boxes A, D, E in vitro. We also demonstrated through ChIP assay that NF-Y was associated with the oriLyt core region in vivo. CCAAT boxes B and C, although did not bind to NF-Y in EMSA (Figure 6), contributed to replication activity of the left oriLyt as demonstrated by the site-directed and the transposon-mediated mutagenesis study (Figure 1 and Figure 4). These data, together with the result using the dominant-negative expression plasmid of NF-Y (Figure 8), suggested the involvement of other factor(s) in mediating the activity of CCAAT boxes B and C.
NF-Y is a ubiquitous, histone-like transcription factor, and each of its three subunits displays highly conserved domains. The NF-YA protein can be divided into subunit association and DNA contacting subdomains. Both conserved domains of NF-YB and NF-YC contain putative histone fold motifs. The tight association between NF-YB and NF-YC is a prerequisite for NF-YA binding and sequence-specific DNA interactions. The CCAAT pentanucleotide is absolutely necessary for NF-Y binding to DNA sequence, although additional flanking sequences are also required. NF-Y binding sites exist in a large amount of eukaryotic promoter regions, and they are frequently located within 50–80 bp of transcriptional initiation sites, or in more distal enhancer control regions of eukaryotic genes. NF-Y is required for the expression of many proteins. It binds to DNA prior to other transcription factors to pre-set the promoter architecture for other regulatory proteins to access and it may also recruit transcription factors by carrying them to promoter regions (Currie, 1998; Dorn et al., 1987; Hooft van Huijsduijnen et al., 1987; Li et al., 1998; Mantovani, 1998; Sinha et al., 1996). However, most studies have focused on NF-Y’s involvement in regulation of gene expression, and few have examined NF-Y’s role in DNA replication.
Up till now, limited research has been carried out on functions of NF-Y in viral infection. It has been reported that NF-Y could bind to vertebrate viral gene promoters and regulate their expression (Borestrom et al., 2003; Lu and Yen, 1996; Yu et al., 2005). To our knowledge, our study is the first to show that NF-Y binds to an origin of DNA replication in vitro and in vivo, and our functional assay employing the dominant negative expression plasmid of NF-Y further demonstrated that NF-Y plays an important role in MHV-68 lytic genome replication. Our finding broadens the understanding of NF-Y’s role in DNA replication and viral infection. We hypothesize that MHV-68 and possibly other gammaherpesviruses may utilize this ubiquitous cellular transcription factor to pre-set the architecture of oriLyt and to recruit viral or other cellular factors to facilitate efficient lytic DNA replication. Further investigation of the detailed mechanisms governing NF-Y’s role in MHV-68 lytic genome replication will provide new insights into herpesvirus lytic replication as well as functions of the cellular transcription factor NF-Y.
METERIALS AND METHODS
Cell culture and virus
All cells were cultured at 37°C in the presence of 5% CO2. 293T cells and BHK-21 cells were grown and maintained in Dulbecco’s modified Eagle’s medium (Gibco) containing 10% fetal bovine serum (Hyclone) and antibiotics (50 U of penicillin and 50 µg of streptomycin per ml). S11E, which is a mouse B cell lymphoma line harboring latent MHV68 genome, was grown in RPMI 1640 medium containing 10% FBS and antibiotics.
MHV-68 was propagated by infecting BHK-21 cells at a multiplicity of infection (MOI) of 0.05 plaque forming unit (PFU)/cell. Viral titers were measured by standard plaque assay.
Plasmid construction
Plasmid pMOL was constructed by cloning a 1.2-kb fragment (NC_001826, nt. 25,695–26,883, amplified by PCR) of MHV-68 DNA into pGEM-T vector (Promega). The internal deletion mutants of pMOL were generated by using a PCR-based mutagenesis system. In brief, a pair of oligonucleotides (one of them was phosphorylated) towards opposite directions were used in a high-fidelity PCR with pMOL plasmid as template. After PCR, the template DNA was removed by digestion with DpnI. The DpnI-digested PCR products were self-ligated and used to transform E.coli competent cells. Site-directed mutagenesis of pMOL was carried out similarly to the internal deletion method, except that one primer contains the desired nucleotide mutation(s). All deletion and site-directed mutations were confirmed by sequencing.
The murine NF-Ys subunits were cloned from S11E cell’s cDNA by PCR with the following primers: NF-YA, 5’-GCG AAT TCC CAT GGA GCA GTA TAC GAC AAA C-3’ and 5’-CAG AGA TCT CCT GTG GTT AGG AAA CTC GGA-3’; NF-YB, 5’-GAC GAA TTC TCA TGA CAA TGG ACG GCG AC-3’ and 5’-CAG AGA TCT GAC TCT CTC CAC ACA AGC CCG-3’; NF-YC, 5’-GAC GAA TTC AAA TGT CCA CAG AAG GAG GGT-3’ and 5’-CAG AGA TCT CAG CTC AGG CCC TCA GTC TC-3’. PCR products were cloned into pSG-Flag vector (pSG-5 vector from Stratagene with a linker containing FlAG epitope sequence inserted into the EcoRI site) to generate the three NF-Y expressing plasmids, which were named pSGFlag-NFYA, pSGFlag-NFYB and pSGFlag-NFYC respectively. Expression plasmid for dominant negative mutant of NF-YA (Δ4NF-YA13m29) was kindly provided by Dr. Roberto Mantovani (Universita di Milano, Milan, Italy) (Mantovani et al., 1994).
Generation and in vitro selection of MHV-68 minimal right oriLyt mutant library
A 1.25-kb region corresponding to nt. 100,724–101,975 in the right end of the MHV-68 genome, which functions as minimal oriLyt (Deng et al., 2004), was used for high-density mutagenesis and functional fine mapping study. A plasmid, pMOΔ16 (Deng et al., 2004), containing this minimal oriLyt was randomly mutagenized using Not I-mini-Mu transposons by Mutation Generation Kit (Finnzymes, Finland) per manufacturer’s protocols. Subsequently, the transposon body was removed by restriction digestion using NotI enzyme. This resulted in a library of mutants having a 15-nucleotide sequence, 5’-NNN NTG CGG CCG CA-3’, (N denotes duplicated target nucleotides at the site of Mu insertion), inserted randomly in the oriLyt region.
To identify the cis-elements essential for the oriLyt replication, 0.8 pmol (or 3.2 pmol) of the oriLyt mutant plasmids were transfected into 293T cells in 10 cm dishes with Lipofectamine Plus (Invitrogen), and subjected to replication assay.
Genetic foot-printing analysis of non-selected and selected right oriLyt mutant library
In order to identify the location of 15-bp insertion in the non-selected (input) and in vitro selected oriLyt mutant library, a modified genetic foot-printing method was performed. Briefly, the mutated oriLyt region was PCR amplified from input and in vitro selected (Dpn I digested) mutant libraries with primers Lac forward (5’-CTG CGC AAC TGT TGG GAA-3’) and M13 reverse (5’-CAG GAA ACA GCT ATG AC-3’). Fifty nanogram of column purified PCR product was subsequently used for a second round of PCR using a fluorescently labeled Not-Mini primer (5’VIC-TGC GGC CGC A-3’) and one of the following right oriLyt specific primers: ORI-1 (5’-GGG AGC CAA AGC GAG GAG CA-3’), ORI-2 (5’-GGT CCC CAT ATT GAA TAT ACC T-3’), ORI-3 (5’-GGA CCA ATC ACC AAC TTG A-3’), and ORI-4 (5’-GGT TTG CGG TTA GAC CAG GCA-3’). The fluorescently labeled PCR products were run on an ABI-3730xl DNA analyzer and data were analyzed using ABI GeneMapper software program. The location of each 15-bp insertions in the oriLyt region was precisely determined based on the genome position of each right oriLyt specific primer that was used for the genetic foot-printing PCR.
De novo infection-replication assay and Southern blot
To test the replication capability of oriLyt containing constructs during de novo infection, 3 µg plasmids were transfected into 293T cells in six-well plates with calcium phosphate. Twenty-four hrs post-transfection, cells were infected with wild type MHV-68 at a MOI of 0.05 to provide the trans-factors required for DNA replication. When >95% cells showed CPE at 72–96 hrs post-infection, cells were harvested and total cellular DNA was extracted and prepared for Southern blot analysis.
One-twelfth of each total cellular DNA was digested overnight with DpnI and a unique restriction enzyme (PstI for pMOL and its deletion plasmids, NdeI for pMOΔ16 and its deletion plasmids). For Southern blot analysis, the digested samples and molecular size markers were run on a 0.8% agarose gel in 1×TAE buffer. The gel was treated with 0.25 M HCl, followed by alkaline denaturation and neutralization. DNA was transferred onto a Hybond-N+ membrane (Amersham Pharmacia) via capillary transfer in 10×SSC buffer and immobilized by UV-crosslinking. Southern blot was carried out with DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) with a probe against the pGEM-T vector.
Nuclear extract preparation
293T cells in 10 cm plate were co-transfected with 4 µg each of pSG-Flag-YA, pSG-Flag-YB and pSG-Flag-YC. Twenty-four hrs post-transfection, cells were harvested and nuclear extracts were prepared using a modified method described by Edgar Schreiber (Schreiber et al., 1989). Briefly, cells from one 10 cm plate were resuspended in 1 ml buffer A (10 mM Hepes pH7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM PMSF), and swelled on ice for 15 min. After 62.5 µl of 10% Nonidet NP-40 (Fluka) was added, the tube was vigorously vortexed for 10 seconds and the solution was passed through a 22G needle for 10 times. The homogenate was centrifuged for 10 min at 4,000g, 4°C. The nuclear pellet was then resuspended in 100 µl of buffer C (20 mM Hepes pH7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 20% glycerol) in the presence of protease inhibitor cocktail tablets (Roche). Cellular debris was removed by centrifuge and nuclear extracts were stored in aliquots at −70°C.
Electrophoretic mobility shift assay (EMSA)
Oligonucleotides were prepared as described in the instruction for Biotin 3’ End DNA Labeling Kit (Pierce). In DNA-protein binding reaction, 3 µl of nuclear extract was incubated in a 20 µl total volume, containing 10 mM Tris-HCl pH7.5, 10 mM NaCl, 0.1 mM DTT, 1 mM EDTA, 5% glycerol, and 1 µg poly(dI-dC). After 10 min incubation on ice, 50 fmol biotin-labeled oligonucleotides were added, and incubated at room temperature for another 20 min. For supershift assay, 1 µl of anti-FLAG antibody (Sigma) was added before incubation with the biotin-labeled oligonucleotides. For competition assay, 200-fold amount of unlabeled oligonucleotides were added. After incubation, 5 µl of 20% (w/v) Ficoll was added to each reaction, and the reactions were loaded on a 5% native polyacrylamide gel in 1×TGE buffer. After electrophoresis, DNA was transferred onto Hybond-N+ membrane and immobilized by UV-crosslinking. Biotin-labeled oligonucleotides were detected according to the method described in the Phototope-Star Chemiluminescent Detection Kit (New England Biolabs).
Oligonucleotides used in the EMSA assay are as follows. CCAAT-boxA, 5’-GCT CAC ATC CTA TTG GCC TGT CAA AAA-3’; BoxB, 5’-CTG TCA AAA AAT CAA TCA AGA AAT CCG-3’; BoxC, 5’-CCG ACC AAG ACA TTG GAT AGT GCC AAA-3’; BoxD, 5’-CTA GCC TGC TCA TTG GCC CAG AGC AAA-3’; BoxE, 5’-AGA GCA AAC AAC CAA TCA GAT GGG AGG-3’. A 22-bp oligonucleotide encompassing the Y box of MHCII E alpha promoter, 5’-ATT TTT CTG ATT GGT TAA AAG T-3’ (Dorn et al., 1987), was used as a cold competitor for NF-Y binding.
Western blotting analysis
Approximately 5×106 cells were pelleted by centrifugation and lysed in 500 µl of loading buffer (0.125 M Tris-HCl pH 6.8, 2% (w/v) SDS, 10% glycerol, 2% β-mercaptoethanol, 0.4 M DTT, 0.05% Bromophenol blue). The cell lysate was boiled for 5 min, and 10 µl of lysate was subjected to gel electrophoresis on a 10% SDS-polyacrylamide gel. Proteins were electrotransferred onto PVDF membrane (Millipore) using a Bio-Rad apparatus. Blocking was performed in phosphate buffered saline plus 0.1% Tween-20 and 5% nonfat powdered milk. To detect the protein(s) of interest, the membrane was incubated with a primary antibody against FLAG epitope overnight. HRP-conjugated goat-anti-mouse immunoglobulin G was used as a secondary antibody. Bands were visualized by incubation with Immobilon Western Chemiluminescent HRP substrate (Millipore) and exposed to an X-ray film (Kodak).
Chromatin immuno-precipitation (ChIP)
2×106 293T cells were transfected with pSGFlag-NFYA/B/C, and 24 hrs later, cells were infected with wild type MHV-68 at a MOI of 5. Twelve hrs post-infection, 2×106 cells were harvested and washed twice with 1×PBS, and proteins were cross-linked to DNA by adding formaldehyde to a final concentration of 1% following by incubation for 15 min at room temperature. Nuclei were extracted, lysed in 450 µl of SDS lysis buffer (50 mM Tris pH 8.1, 1% SDS, 10 mM EDTA, and protease inhibitor cocktail) for 10 min on ice, and sonicated on ice to fragmentate the genomic DNA. The lysate was centrifuged to remove cellular debris. For chromatin immuno-precipitation, 200 µl of supernatant was diluted in 1800 µl of ChIP dilution buffer (16.7 mM Tris-HCl, pH8.1, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, and the protease inhibitor cocktail), and the samples were incubated with 5 µl of monoclonal anti-FLAG antibody; a negative control was performed with no antibody. Immune complexes were precipitated by adding 40 µl of protein G-sepharose beads (Upstate), and centrifuged at low speed. Sepharose beads were sequentially washed once with low salt immune complex wash buffer, high salt immune complex wash buffer and LiCl buffer, and washed twice with TE pH 8.0. DNA/protein complexes were eluted from the beads by incubating in 250 µl of elution buffer (1% SDS, 0.1 M NaHCO3) for 15 min twice. Twenty microliter of 5 M NaCl was added to the combined eluates and DNA/proteins cross-links were reversed by incubation at 65°C for 4 hrs. Following treatment with proteinase K, DNA was extracted with phenol/chloroform and precipitated with ethanol. Primer pairs used to amplify a region of interest within the left oriLyt, the RTA promoter and the human beta-actin coding region are: MOL6F, 5’-GCT ATG TTT GAC TTT TCG CTG TTT CG-3’, and MOL7.3R, 5’-AAG GGG ATT TCC AGG TAG AGG GTC TTC-3’; 5’-CTG GCA ACC ACC ACC TTC AC-3’, and 5’-GCA GAA ATT CCC TCG TAG TGC-3’; and 5’-GGA CTT CGA GCA AGA GAT GG-3’, and 5’-AGC ACT GTG TTG GCG TAC AG-3’, respectively. PCR products were analyzed on a 2% agarose gel.
ACKNOWLEDGMENTS
We thank Dr. Roberto Mantovani for kindly providing the expression plasmid Δ4NF-YA13m29, and members of the Deng and Sun laboratory for kind help and discussion. This work was supported by National Science Foundation grant 30670076, NIH grant DE15612 and International Fogarty Program on AIDS Malignancies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adler H, Steer B, Freimuller K, Haas J. Murine gammaherpesvirus 68 contains two functional lytic origins of replication. J Virol. 2007;81(13):7300–7305. doi: 10.1128/JVI.02406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AuCoin DP, Colletti KS, Xu Y, Cei SA, Pari GS. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J Virol. 2002;76(15):7890–7896. doi: 10.1128/JVI.76.15.7890-7896.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borestrom C, Zetterberg H, Liff K, Rymo L. Functional interaction of nuclear factor y and sp1 is required for activation of the epstein-barr virus C promoter. J Virol. 2003;77(2):821–829. doi: 10.1128/JVI.77.2.821-829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CM, Medveczky MM, Lund T, Medveczky PG. The terminal repeats and latency-associated nuclear antigen of herpesvirus saimiri are essential for episomal persistence of the viral genome. J Gen Virol. 2002;83(Pt 9):2269–2278. doi: 10.1099/0022-1317-83-9-2269. [DOI] [PubMed] [Google Scholar]
- Currie RA. Biochemical characterization of the NF-Y transcription factor complex during B lymphocyte development. J Biol Chem. 1998;273(29):18220–18229. doi: 10.1074/jbc.273.29.18220. [DOI] [PubMed] [Google Scholar]
- Deng H, Chu JT, Park NH, Sun R. Identification of cis sequences required for lytic DNA replication and packaging of murine gammaherpesvirus 68. J Virol. 2004;78(17):9123–9131. doi: 10.1128/JVI.78.17.9123-9131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A, Durand B, Marfing C, Le Meur M, Benoist C, Mathis D. Conserved major histocompatibility complex class II boxes--X and Y--are transcriptional control elements and specifically bind nuclear proteins. Proc Natl Acad Sci U S A. 1987;84(17):6249–6253. doi: 10.1073/pnas.84.17.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi B, Dutia BM, Roberts KL, Garcia-Ramirez JJ, Dickinson P, Stewart JP, Ghazal P, Roy DJ, Nash AA. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J Gen Virol. 2003;84(Pt 1):99–109. doi: 10.1099/vir.0.18639-0. [DOI] [PubMed] [Google Scholar]
- Efstathiou S, Ho YM, Hall S, Styles CJ, Scott SD, Gompels UA. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J Gen Virol. 1990;71(Pt 6):1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- Farina A, Manni I, Fontemaggi G, Tiainen M, Cenciarelli C, Bellorini M, Mantovani R, Sacchi A, Piaggio G. Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene. 1999;18(18):2818–2827. doi: 10.1038/sj.onc.1202472. [DOI] [PubMed] [Google Scholar]
- Fixman ED, Hayward GS, Hayward SD. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66(8):5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixman ED, Hayward GS, Hayward SD. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69(5):2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55(3):427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- Hooft van Huijsduijnen RA, Bollekens J, Dorn A, Benoist C, Mathis D. Properties of a CCAAT box-binding protein. Nucleic Acids Res. 1987;15(18):7265–7282. doi: 10.1093/nar/15.18.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Renne R. Characterization of the minimal replicator of Kaposi's sarcoma-associated herpesvirus latent origin. J Virol. 2005;79(4):2637–2642. doi: 10.1128/JVI.79.4.2637-2642.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E, Rickinson AB. Epstein-Barr virus and its replication. In: Howley DMKaPM., editor. Fields virology. 4 ed. Vol. 2. Philadelphia, Pa: Lippincott Williams & Wilkins; 2001. pp. 2511–2627. [Google Scholar]
- Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko VV, Nakatani Y, Wolffe AP. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylationresponsive transcription from the Xenopus hsp70 promoter in vivo. Embo J. 1998;17(21):6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati C, di Silvio A, Ottolenghi S, Mantovani R. NF-Y binding to twin CCAAT boxes: role of Q-rich domains and histone fold helices. J Mol Biol. 1999;285(4):1441–1455. doi: 10.1006/jmbi.1998.2384. [DOI] [PubMed] [Google Scholar]
- Lin CL, Li H, Wang Y, Zhu FX, Kudchodkar S, Yuan Y. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J Virol. 2003;77(10):5578–5588. doi: 10.1128/JVI.77.10.5578-5588.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SF, Robinson DR, Miller G, Kung HJ. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol. 1999;73(3):1909–1917. doi: 10.1128/jvi.73.3.1909-1917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CC, Yen TS. Activation of the hepatitis B virus S promoter by transcription factor NF-Y via a CCAAT element. Virology. 1996;225(2):387–394. doi: 10.1006/viro.1996.0613. [DOI] [PubMed] [Google Scholar]
- Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26(5):1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R, Li XY, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269(32):20340–20346. [PubMed] [Google Scholar]
- Martinez-Guzman D, Rickabaugh T, Wu TT, Brown H, Cole S, Song MJ, Tong L, Sun R. Transcription program of murine gammaherpesvirus 68. J Virol. 2003;77(19):10488–10503. doi: 10.1128/JVI.77.19.10488-10503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ESJ, Courcelle CT. Cytomegaloviruses and their replication. In: Howley DMKaPM., editor. Fields virology. 4 ed. Vol. 2. Philadelphia, Pa: Lippincott Williams & Wilkins; 2001. pp. 2629–2673. [Google Scholar]
- Nash AA, Dutia BM, Stewart JP, Davison AJ. Natural history of murine gamma-herpesvirus infection. Philos Trans R Soc Lond B Biol Sci. 2001;356(1408):569–579. doi: 10.1098/rstb.2000.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Chittenden T, Levine AJ. Identification of cellular factors that bind specifically to the Epstein-Barr virus origin of DNA replication. J Virol. 1991;65(1):514–519. doi: 10.1128/jvi.65.1.514-519.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pari GS, AuCoin D, Colletti K, Cei SA, Kirchoff V, Wong SW. Identification of the rhesus macaque rhadinovirus lytic origin of DNA replication. J Virol. 2001;75(23):11401–11407. doi: 10.1128/JVI.75.23.11401-11407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Knipe DM. Herpes simplex viruses and their replication. In: Howley DMKaPM., editor. Fields virology. 4 ed. Vol. 2. Philadelphia, Pa: Lippincott Williams & Wilkins; 2001. pp. 2399–2460. [Google Scholar]
- Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. Embo J. 2001;20(16):4588–4602. doi: 10.1093/emboj/20.16.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989;17(15):6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simas JP, Efstathiou S. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 1998;6(7):276–282. doi: 10.1016/s0966-842x(98)01306-7. [DOI] [PubMed] [Google Scholar]
- Sinha S, Maity SN, Seldin MF, de Crombrugghe B. Chromosomal assignment and tissue expression of CBF-C/NFY-C, the third subunit of the mammalian CCAAT-binding factor. Genomics. 1996;37(2):260–263. doi: 10.1006/geno.1996.0555. [DOI] [PubMed] [Google Scholar]
- Stevenson PG. Immune evasion by gamma-herpesviruses. Curr Opin Immunol. 2004;16(4):456–462. doi: 10.1016/j.coi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Virgin HWt, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71(8):5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Dong L, Saville B, Safe S. Transcriptional activation of E2F1 gene expression by 17beta-estradiol in MCF-7 cells is regulated by NF-Y-Sp1/estrogen receptor interactions. Mol Endocrinol. 1999;13(8):1373–1387. doi: 10.1210/mend.13.8.0323. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li H, Chan MY, Zhu FX, Lukac DM, Yuan Y. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J Virol. 2004;78(16):8615–8629. doi: 10.1128/JVI.78.16.8615-8629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Rodriguez-Huete A, Pari GS. Evaluation of the lytic origins of replication of Kaposi's sarcoma-associated virus/human herpesvirus 8 in the context of the viral genome. J Virol. 2006;80(19):9905–9909. doi: 10.1128/JVI.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue SA, Griffin BE. Complexities associated with expression of Epstein-Barr virus (EBV) lytic origins of DNA replication. Nucleic Acids Res. 2007;35(10):3391–3406. doi: 10.1093/nar/gkm170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zhu X, Pi W, Ling J, Ko L, Takeda Y, Tuan D. The long terminal repeat (LTR) of ERV-9 human endogenous retrovirus binds to NF-Y in the assembly of an active LTR enhancer complex NF-Y/MZF1/GATA-2. J Biol Chem. 2005;280(42):35184–35194. doi: 10.1074/jbc.M508138200. [DOI] [PubMed] [Google Scholar]