Abstract
Vitamin D serves as a principal modulator of skeletal gene transcription, thus necessitating an understanding of interfaces between the activity of this steroid hormone and regulatory cascades that are functionally linked to the regulation of skeletal genes. Physiological responsiveness requires combinatorial control where coregulatory proteins determine the specificity of biological responsiveness to physiological cues. It is becoming increasingly evident that the regulatory complexes containing the vitamin D receptor are dynamic rather than static. Temporal and spatial modifications in the composition of these complexes provide a mechanism for integrating regulatory signals to support positive or negative control through synergism and antagonism. Compartmentalization of components of vitamin D control in nuclear microenvironments supports the integration of regulatory activities, perhaps by establishing thresholds for protein activity in time frames that are consistent with the execution of regulatory signaling.
1. Introduction
Vitamin D is a physiologically significant mediator of biological control. Vitamin D plays a principal role in control of proliferation and differentiation throughout development and during tissue remodeling in adults. It has been extensively documented that vitamin D has an important function during bone formation and resorption, immune system maturation, and gastrointestinal absorption [1,2]. Equally important, vitamin D has been implicated in regulatory activity associated with the onset and progression of cancer. Vitamin D is linked to signaling cascades that contribute to prostate and colon cancer and leukemogenesis. Therefore, beyond physiological control, vitamin D analogs can be effective chemotherapeutic agents.
Vitamin D regulation is principally mediated through modulation of transcription. Vitamin D3 binds to the vitamin D receptor (VDR), which then interacts with specific elements located within the regulatory regions of target genes. Combinatorial and context-dependent protein-protein interactions with other transcription factors or cofactors bound at a specific promoter may further modify transcription. Here, physiological responsiveness requires that co-regulatory proteins determine specificity of biological responsiveness to regulatory cues.
It is becoming increasingly evident that organization and assembly of VDR-regulatory complexes are dynamic rather than static [3]. Modifications in the composition of these regulatory complexes provide a mechanism for integrating regulatory signals to support positive and negative control through synergism and antagonism.
In addition to the genomic effects of vitamin D, there is increasing insight into the mechanisms that are related to rapid non-genomic actions [4]. These genomic and non-genomic influences of vitamin D need not be mutually exclusive. In fact, rapid non-genomic and longer term actions may be synergistic. The challenge then is to define pathways where the intersection of genomic and non-genomic actions occurs.
This article will focus on genomic actions of vitamin D3 and review mechanisms that support the organization, assembly and activities of regulatory complexes that determine the temporal and spatial parameters of vitamin D control. We will address biological control of skeletogenesis by vitamin D3.
2. Components of regulatory complexes
The role that vitamin D (1α,25-dihydroxy vitamin D3) plays in bone metabolism provides a paradigm for understanding molecular mechanisms that operate in vitamin D action. Vitamin D directly regulates the expression of genes that support bone formation during development and bone remodeling throughout life [1]. Vitamin D exerts its genomic effects through the vitamin D receptor (VDR) which is a member of the superfamily of nuclear receptors [3,5]. As in other nuclear receptors, binding of the ligand induces conformational changes in the C-terminal ligand binding domain (LBD) of the VDR. The changes establish competency for VDR interaction with coactivators of the p160/SRC family, including SRC-1/NCoA-1, SRC-2/NCoA-2/GRIP/TIF2, and SRC-3/ACTR. These complexes are critical for transcriptional activation [1,3,5]. p160/SRC coactivators form high molecular weight complexes by interacting with other coactivator proteins including p300, its related homologue CBP, and P/CAF [6].
Moreover, p160/SRC coactivators have been shown to recruit CBP/p300 and P/CAF to ligand-bound nuclear receptors. Multiprotein complexes containing different activities are functionally linked to ligand-dependent transcriptional regulation [3]. Coactivators such as SRC-3/ACTR, SRC-1/NCoA-1, CBP/p300 and P/CAF contain intrinsic histone acetyl transferase (HAT) activity. Therefore, protein complexes including independent HAT activities can be recruited to gene promoters by nuclear receptors in a ligand-dependent manner [3]. Once bound to these promoters, the HAT activities contribute to chromatin remodeling events that increase access of additional regulatory factors to their cognate elements [7].
The multisubunit DRIP (VDR-Interacting Protein) complex also binds to VDR in response to the ligand vitamin D [8,9]. This interaction occurs through the LBD of VDR in the same manner as the p160/SRC coactivators, resulting in transcriptional enhancement [10]. In contrast to p160/SRC coactivators, DRIP is devoid of HAT and other chromatin remodeling activities and interacts with nuclear receptors through a single subunit designated DRIP205, which anchors other subunits to the receptor LBD. Several of these subunits are also present in the Mediator complex, which interacts with the C-terminal domain (CTD) of RNA polymerase II, forming the holoenzyme complex [11]. Therefore, the DRIP complex appears to function as a transcriptional coactivator by forming a molecular bridge between the VDR and the basal transcription machinery.
Recently, a novel ATP-dependent chromatin remodeling complex that binds to VDR, termed WINAC (for WSTF [Williams Syndrome Transcription Factor]-including nucleosome assembly complex), has been reported [12]. WINAC shares components with two other chromatin-remodeling complexes, SWI/SNF and ISWI, and has been proposed to mediate recruitment of unliganded VDR to target genes. Nevertheless, subsequent interaction of the targeted VDR with transcriptional coregulators requires the presence of vitamin D. This complex has been reported to be involved in transcriptional repression [13] and in controlling DNA replication [12].
In the last few years various investigators have shown that coactivator complexes including p160/SRC and DRIP are recruited to steroid hormone-regulated genes by nuclear receptor in a sequential and mutually exclusive manner [14–17]. The ordered association of transcriptional regulators exhibits binding kinetics with periods of 40 to 60 minutes. These results provided the basis for a model (Figure 1) in which cyclical association of different coactivator complexes reflects the dynamics of the transcription activation process of nuclear receptor-regulated genes [5,18]. Alternatively, recent reports indicate that occupancy at the target gene regulatory regions by nuclear receptor-associated coactivator complexes may also occur gradually and at a significantly lower rate [19,20]. Thus, it has been shown that during keratinocyte differentiation, there is a specific utilization of p160/SRC or DRIP205/Mediator coactivator complexes to regulate vitamin D-dependent genes [20]. The proposed model indicates that both coactivator complexes have important roles during early stages of keratinocyte differentiation, but a subsequent decrease in major DRIP/Mediator components leads to a predominant role for p160/SRC in the later stages of differentiation.
Figure 1.
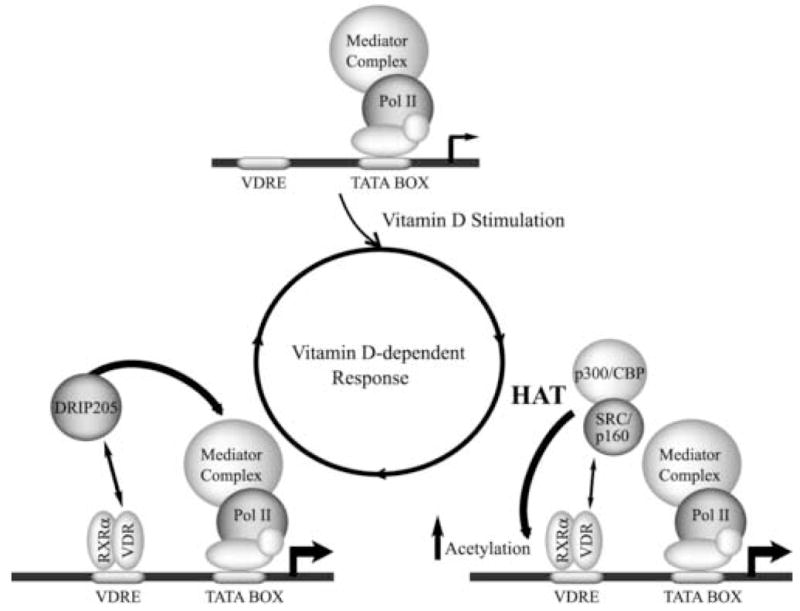
Cyclical and mutually exclusive VDR-mediated recruitment of SRC/p160 and DRIP205 coactivators to vitamin D-target genes. The arrowhead indicates the transcription start site and represents the intensity of the transcription process. VDRE = vitamin D-responsive element, RNA pol II = RNA polymerase II, HAT= histone acetyl transferase activity.
3. Vitamin D-mediated gene expression within the three dimensional context of nuclear structure
Evidence is accumulating that the architectural organization of nucleic acids and regulatory proteins within the nucleus supports functional interrelationships between nuclear structure and gene expression. There is increasing acceptance that components of nuclear structure are functionally linked to the organization and sorting of regulatory information in a manner that permits utilization (reviewed by [21]). The primary level of organization, the representation and ordering of genes and promoter elements, provides alternatives for physiological control. The molecular organization of regulatory elements, the overlap of regulatory sequences within promoter domains, and the multipartite composition of regulatory complexes increase options for responsiveness. Chromatin structure and nucleosome organization reduce distances between regulatory sequences, facilitate cross talk between promoter elements, and render elements competent for interactions with positive and negative regulatory factors. The components of higher order nuclear architecture, including nuclear pores [22], the nuclear matrix, and subnuclear domains, contribute to the subnuclear distribution and activities of genes and regulatory factors [21,23]. Compartmentalization of regulatory complexes is illustrated by focal organization of PML bodies [24], Runx bodies [25,26], the nucleolus [27], and chromosomes [28], as well as by the punctate intranuclear distribution of sites for replication [29], DNA repair [30], transcription [31], and the processing of gene transcripts [32–34]. There is emerging recognition that nuclear structure and function are casually related. The bone-specific OC gene and skeletal-restricted Runx2 transcription factor serve as examples of obligatory relationships between nuclear structure and vitamin D-mediated physiological control of skeletal gene expression [35,36]. It appears that there are similar relationships between nuclear organization and other vitamin D-responsive genes (e.g., osteopontin and 24-hydroxylase). However, we will largely confine our consideration of nuclear structure-gene expression relationships to the OC gene.
3.1. The osteocalcin gene
The rat OC gene encodes a 10 kDa bone-specific protein that is induced in osteoblasts with the onset of mineralization at late stages of differentiation [37]. Modulation of OC gene expression during bone formation and remodeling requires physiologically responsive accessibility of proximal and upstream promoter sequences to regulatory and coregulatory proteins, as well as protein-protein interactions that integrate independent promoter domains [38]. The chromatin organization of the OC gene illustrates dynamic remodeling of a promoter to accommodate requirements for phenotype-related developmental and vitamin D-responsive activity [35].
Transcription of the OC gene is controlled by modularly organized basal and hormone-responsive promoter elements (see Figure 2A), located within two DNase I-hypersensitive sites (Distal site, positions −600 to −400; proximal site, positions −170 to −70) that are only nuclease accessible in bone-derived cells expressing this gene [38]. A key regulatory element that controls OC gene expression is recognized by the VDR complex upon ligand stimulation. This vitamin D responsive element (VDRE) is located in the distal region (Figure 2A) of the OC promoter (positions −465 to −437) and functions as an enhancer to increase OC gene transcription [35]. Another key regulator of OC gene expression is the nuclear matrix-associated transcription factor Runx2, a member of the Runt homology family of proteins which has been shown to contribute to the control of skeletal gene expression [36]. Runx2 proteins serve as a scaffold for the assembly and organization of coregulatory proteins that mediate biochemical and architectural control of promoter activity. The rat OC gene promoter contains three recognition sites for Runx2 interactions, site A (−605 to −595), site B (−438 to −430), and site C (−138 to −130). Mutation of all three Runx2 sites results in significantly reduced OC expression in bone-derived cells [39]. The retention of a nucleosome between the proximal and upstream enhancer domains reduces the distance between the basal regulatory elements and the VDRE and supports a promoter configuration that is conducive to protein-protein interactions between VDR-associated proteins and components of the RNA polymerase II-bound complex (Figure 2B). Interaction of the VDR at the distal promoter region of the OC gene requires nucleosomal remodeling [40,41].
Figure 2.
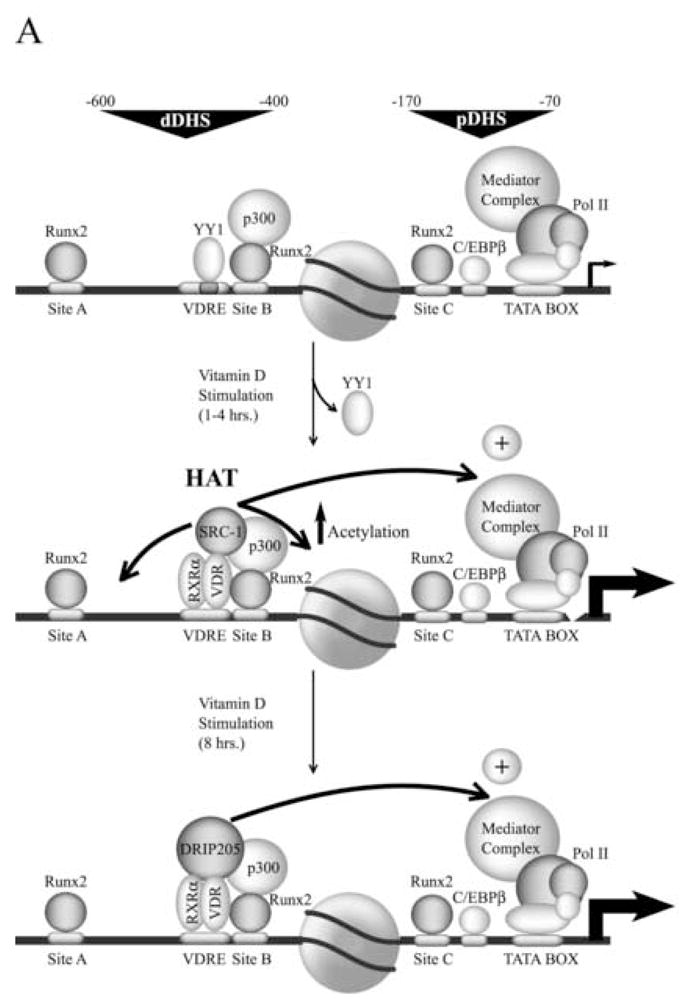
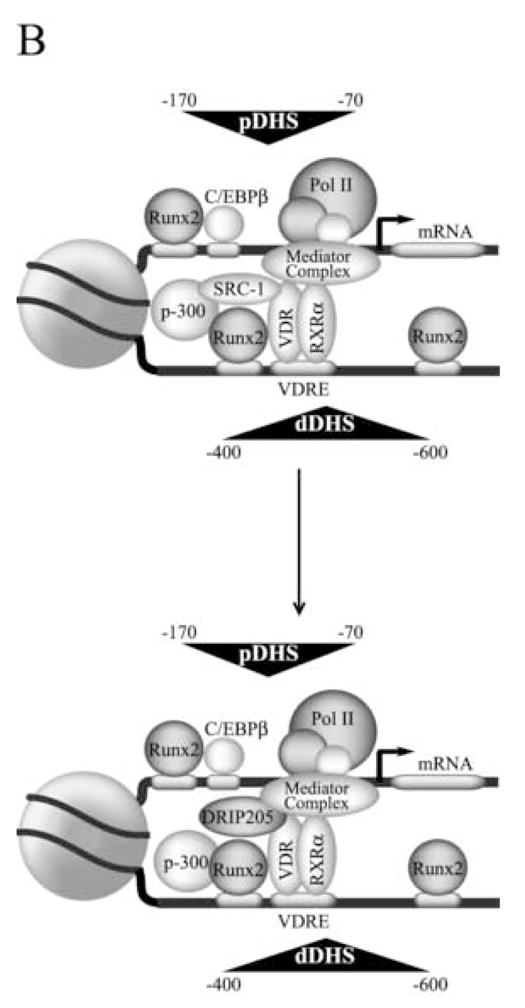
A) Schematic representation of the rat osteocalcin gene promoter transcribing at basal levels (Top panel) or enhanced by vitamin D (middle and lower panels). The circle in the middle represents a positioned nucleosome flanked by a distal and proximal DNase I hypersensitive sites (dDHS and pDHS, respectively). The different transcription factors bound to the promoter are indicated. The stimulatory effect of the VDR-bound coactivators on the general transcription machinery is represented by arrows. B) Proposed model for the three dimensional organization of the osteocalcin gene promoter. The positioned nucleosome facilitates DNA bending and the functional interaction between distal and proximal promoter regulatory elements that is mediated by the associated factors. For an explanation of the other symbols see figure legend 1.
We have recently shown that within the OC gene promoter context there is a tight functional relationship between Runx2 and the vitamin D -dependent pathway [42]. Runx2 and VDR are components of the same nuclear complexes, colocalize at punctate foci within the nucleus of osteoblastic cells, and interact directly in protein-protein binding assays in vitro [42]. As with Runx2, the VDR has been shown to be recruited to the nuclear matrix fraction [43,44], therefore, raising the possibility that both proteins form nuclear matrix-bound regulatory complexes in bone-derived cells. Additionally, mutation of the distal Runx2 sites A and B (which flank the VDRE, see Figure 2A) abolishes vitamin D-enhanced OC promoter activity [42]. In contrast to most nuclear receptors, the VDR does not contain an AF-1 transactivation domain at the N-terminal end and thus is unable to interact with coactivators through this domain [3]. Therefore, Runx2 plays a key role in the vitamin D-dependent stimulation of the OC gene promoter in osteoblastic cells by directly stabilizing binding of the VDR to the VDRE. Runx2 also allows recruitment of the coactivator p300 to the OC promoter (Figure 2A), which results in upregulation of both basal and vitamin D-enhanced OC gene transcription [45]. Based on these results, we have postulated that Runx2-mediated recruitment of p300 may facilitate the subsequent interaction of p300 with the VDR upon ligand stimulation [42].
The rate of recruitment of p160/SCR-1 and DRIP coactivators to the OC gene in response to vitamin D has recently been studied. It has been found that the VDR and SRC-1 rapidly and stably interact with the distal region of the OC promoter encompassing the VDRE (Figure 2A). The interaction of SRC-1 and VDR directly correlates with vitamin D-mediated transcriptional enhancement of the OC gene, increased association of the RNA polymerase complex and vitamin D-stimulated histone H4 acetylation [46,47]. Interestingly, DRIP205 was found to bind to the OC promoter only after several hours of continuous treatment with vitamin D, concomitant with release of SRC-1 (see Figure 2A). Based on these results it has been postulated that this preferential recruitment of SRC-1 to the OC gene promoter is based on the specific distribution of regulatory elements at the distal region of the promoter. This organization may lead to the formation of a stable complex at the distal region that includes Runx2, p300, VDR, and SRC-1. Once established, this complex may directly stimulate the basal transcription machinery bound to an OC promoter actively engaged in transcription. The general relevance of vitamin D-mediated chromatin-based mechanisms of promoter activity, accessibility and cross-talk between regulatory domains, is illustrated by the vitamin D-responsiveness of the osteopontin and 24-hydroxylase genes.
3.2. The 25-hydroxy vitamin D3 24-hydroxylase gene
One of the most pronounced effects of vitamin D is to induce the expression of the 25-hydroxy vitamin D3 24-hydroxylase [24(OH)ase] gene, which encodes an enzyme that metabolizes vitamin D. It has been generally believed that vitamin D-mediated transcription of the 24(OH)ase gene is controlled by VDREs located in the proximal promoter region [17,48,49]. However, recent reports indicate that additional upstream VDR binding regions (see Figure 3A) may be also contributing to full vitamin D-responsiveness by the 24(OH)ase gene in osteoblastic cells [50].
Figure 3.
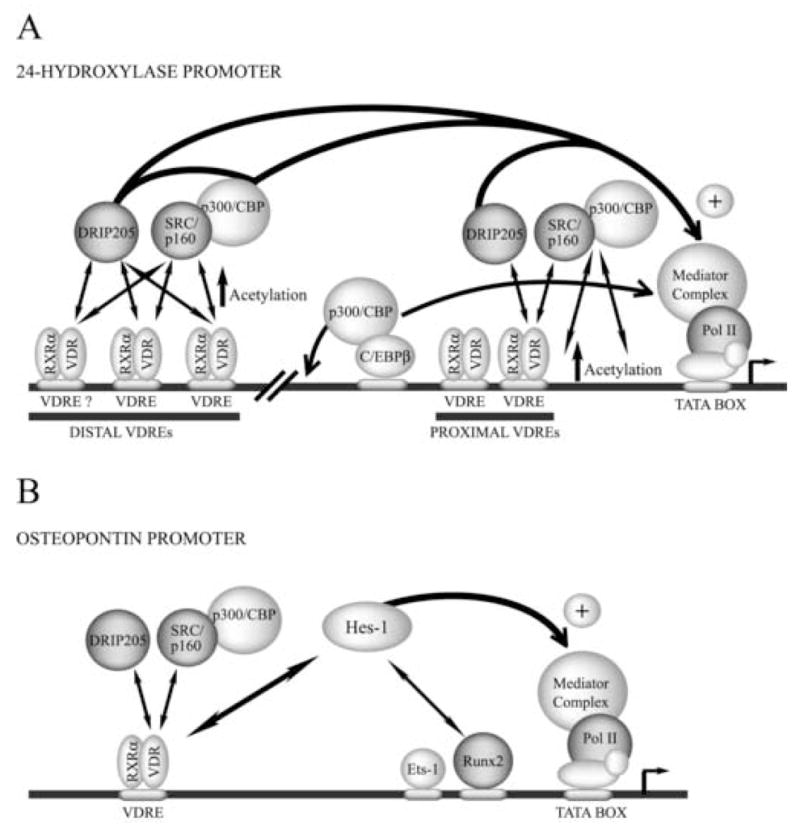
A) Schematic diagram of the 24-hydroxylase gene promoter and vitamin D-dependent transcription. B) Vitamin D-dependent up-regulation of the osteopontin gene. See figure legends 1 and 2 for details of the symbols.
Among the critical elements regulating vitamin D-mediated up-regulation of 24(OH)ase is a C/EBP binding site that has been identified within the proximal promoter region of the rat 24(OH)ase gene (−395 to −388). The C/EBPβ transcription factor binds to this element in osteoblastic cells and recruits the coactivator p300/CBP. This complex then cooperates with the VDR bound to the proximal VDRE site to increase 24(OH)ase gene transcription [49]. Based on these results it has been postulated that crosstalk between the C/EBP family of transcription factors and the VDR may be operating during vitamin D-induced 24(OH)ase gene transcription (Figure 3A).
Other recent studies have established that vitamin D induces a rapid and cyclical association of the VDR/RXR heterodimer with the proximal mouse 24(OH)ase gene promoter in osteoblastic cells [17]. Vitamin D treatment also induces a rapid recruitment of coactivators such as p160/SRC and p300/CBP, which leads to acetylation of histone H4. DRIP205/Mediator is also recruited to the proximal promoter region concomitantly with the interaction of RNA polymerase II. Together, these results support a model in which highly dynamic association of the VDR with chromatin occurs during vitamin D-dependent induction of the 24(OH)ase gene in osteoblasts [17].
Carlberg et al. have also monitored the spatio-temporal regulation of the human 24(OH)ase gene [50]. They have evaluated 25 contiguous genomic regions spanning the first 7.7 kb of the human 24(OH)ase promoter and found that in addition to the proximal VDREs, three further upstream regions are associated with the VDR upon vitamin D stimulation. Interestingly, only two of these regions contain sequences that resemble known VDREs that are transcriptionally responsive to this hormone. The other VDR-associated upstream promoter region does not contain any recognizable classical VDRE that could account for the presence of the VDR protein. However, simultaneous association of the VDR, RXR, p160/SRC and DRIP/Mediator coactivators, as well as RNA polymerase was detected in all four vitamin D-responsive sequences after the addition of the ligand [50]. Remarkably, despite participating in the same process, all four chromatin regions displayed individual vitamin D-dependent patterns of interacting proteins. Based on these results, the authors propose that these upstream vitamin D-responsive regions may have a role in the implementation of gene activation, as they raise their vitamin D-dependent histone H4 acetylation status increases earlier than that of the proximal promoter VDREs [50]. It has also been suggested that the simultaneous communication of the individual promoter regions with the RNA polymerase II complex occurs through a particular three-dimensional organization of the chromatin at the 24(OH)ase promoter. This arrangement could be facilitating close contact between distal and proximal regulatory regions.
3.3. The osteopontin gene
Osteopontin (OP) is an extracellular matrix protein that contains integrin binding motifs that are required for the attachment of osseous cells to the bone-surface. Vitamin D treatment results in increased OP expression in osteoblastic cells as the VDR/RXR heterodimer binds to the OP promoter (−757 to −743) by recognizing a perfect DR3 motif [51]. The OP promoter also contains a Runx-binding site (−136 to −130) which is recognized by the Runx2 factor resulting in transcriptional enhancement [52].
Recent reports indicate that in osteoblastic cells the VDR and Runx2 cooperate to up-regulate OP transcription [52]. Moreover, vitamin D treatment results in increased binding of both Runx2 and the VDR to the OP promoter. Hes-1, a down-regulator of the Notch signaling pathway, was found to form a complex with Runx2 in the nuclei of osteoblastic cells, an interaction that was further increased by treatment of the cells with vitamin D. Together, these results lead to the proposition of a novel mechanism in which three different pathways - - Runx2, vitamin D, and Notch signaling - - intersect at the OP promoter to regulate transcription in osteoblastic cells.
Pike et al. have described that upon vitamin D-treatment of osteoblastic cells, there is a rapid and cyclical association of the VDR with the OP promoter [17]. This increased binding of the VDR parallels vitamin D-mediated transcriptional enhancement of the OP gene and additionally involves cyclical, sequential, and mutually exclusive recruitment of the coactivators p160/SRC, p300/CBP, and DRIP/Mediator. Interestingly and in contrast to the OC and 24(OH)ase genes, p160/SRC-p300/CBP binding does not result in increased histone H4 acetylation. These results further confirm that in osteoblastic cells different promoters are regulated by distinct mechanisms in response to vitamin D.
4. Conclusions and future directions
Vitamin D serves as a principal modulator of skeletal gene transcription, thus necessitating an understanding of interfaces between activity of this steroid hormone with regulatory cascades that are functionally linked to regulation of skeletal genes [35]. There is growing appreciation for the repertoire of factors that influence gene expression for commitment to the osteoblast lineage. It is well documented that sequentially expressed genes support progression of osteoblast differentiation through developmental transition points where responsiveness to phosphorylation-mediated regulatory cascades determine competency for establishing and maintaining the structural and functional properties of bone cells [53,54]. The catalog of promoter elements and cognate regulatory proteins that govern skeletal gene expression offer essential but insufficient insight into mechanisms that are operative in intact cells. Gene promoters serve as regulatory infrastructure by functioning as blueprints for responsiveness to the flow of cellular regulatory signals. However, access to the specific genetic information requires transcriptional control of skeletal genes within the context of the subnuclear organization of nucleic acids and regulatory proteins. Explanations are required for (1) convergence of multiple regulatory signals at promoter sequences; (2) the integration of regulatory information at independent promoter domains; (3) selective utilization of redundant regulatory pathways; (4) thresholds for initiation or down-regulation of transcription with limited intranuclear representation of promoter elements and regulatory factors; (5) mechanisms that render the promoters of cell growth and phenotypic genes competent for protein-DNA and protein-protein interactions in a physiologically responsive manner; (6) the composition, organization, and assembly of sites within the nucleus that support transcription, and (7) the intranuclear trafficking of regulatory proteins to transcriptionally active foci. From a regulatory perspective compartmentalization of components of vitamin D3 control supports the integration of regulatory activities, perhaps by establishing thresholds for protein activity in time frames that are consistent with the execution of regulatory signaling.
Acknowledgments
This work was supported by grants from CONICYT-PBCT ACT044 (to M.M.), NIH PO1 AR48818 (to G.S.S) and DE12528 (to J. B. L.). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. The authors thank Betsy Bronstein for editorial assistance with the preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Christakos S, Dhawan P, Liu Y, Peng X, Porta A. New insights into the mechanisms of vitamin D action. J Cell Biochem. 2003;88:695–705. doi: 10.1002/jcb.10423. [DOI] [PubMed] [Google Scholar]
- 2.van DM, Pols HA, Van Leeuwen JP. Osteoblast differentiation and control by vitamin D and vitamin D metabolites. Curr Pharm Des. 2004;10:2535–2555. doi: 10.2174/1381612043383818. [DOI] [PubMed] [Google Scholar]
- 3.Rachez C, Freedman LP. Mechanisms of gene regulation by vitamin D(3) receptor: a network of coactivator interactions. Gene. 2000;246:9–21. doi: 10.1016/s0378-1119(00)00052-4. [DOI] [PubMed] [Google Scholar]
- 4.Norman AW, Mizwicki MT, Norman DP. Steroid-hormone rapid actions, membrane receptors and a conformational ensemble model. Nat Rev Drug Discov. 2004;3:27–41. doi: 10.1038/nrd1283. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, O’Malley BW. Molecular mechanisms and cellular biology of the steroid receptor coactivator (SRC) family in steroid receptor function. Rev Endocr Metab Disord. 2002;3:185–192. doi: 10.1023/a:1020016208071. [DOI] [PubMed] [Google Scholar]
- 6.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 7.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 8.Rachez C, Suldan Z, Ward J, Chang CP, Burakov D, Erdjument-Bromage H, Tempst P, Freedman LP. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 10.Rachez C, Gamble M, Chang CP, Atkins GB, Lazar MA, Freedman LP. The DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes. Mol Cell Biol. 2000;20:2718–2726. doi: 10.1128/mcb.20.8.2718-2726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa H, Fujiki R, Yoshimura K, Mezaki Y, Uematsu Y, Matsui D, Ogawa S, Unno K, Okubo M, Tokita A, Nakagawa T, Ito T, Ishimi Y, Nagasawa H, Matsumoto T, Yanagisawa J, Kato S. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113:905–917. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- 13.Fujiki R, Kim MS, Sasaki Y, Yoshimura K, Kitagawa H, Kato S. Ligand-induced transrepression by VDR through association of WSTF with acetylated histones. EMBO J. 2005;24:3881–3894. doi: 10.1038/sj.emboj.7600853. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 15.Sharma D, Fondell JD. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc Natl Acad Sci U S A. 2002;99:7934–7939. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burakov D, Crofts LA, Chang CP, Freedman LP. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J Biol Chem. 2002;277:14359–14362. doi: 10.1074/jbc.C200099200. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20:305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- 18.Metivier R, Reid G, Gannon F. Transcription in four dimensions: nuclear receptor-directed initiation of gene expression. EMBO Rep. 2006;7:161–167. doi: 10.1038/sj.embor.7400626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Oda Y, Sihlbom C, Chalkley RJ, Huang L, Rachez C, Chang CP, Burlingame AL, Freedman LP, Bikle DD. Two distinct coactivators, DRIP/mediator and SRC/p160, are differentially involved in vitamin D receptor transactivation during keratinocyte differentiation. Mol Endocrinol. 2003;17:2329–2339. doi: 10.1210/me.2003-0063. [DOI] [PubMed] [Google Scholar]
- 21.Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Montecino M, Stein JL, van Wijnen AJ, Lian JB, Stein GS. The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep. 2005;6:128–133. doi: 10.1038/sj.embor.7400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iborra FJ, Jackson DA, Cook PR. The path of RNA through nuclear pores: apparent entry from the sides into specialized pores [In Process Citation] J Cell Sci. 2000;113(Pt 2):291–302. doi: 10.1242/jcs.113.2.291. [DOI] [PubMed] [Google Scholar]
- 23.Misteli T. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J Cell Sci. 2000;113:1841–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- 24.Dyck JA, Maul GG, Miller WH, Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 25.Zeng C, McNeil S, Pockwinse S, Nickerson JA, Shopland L, Lawrence JB, Penman S, Hiebert SW, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaidi SK, Javed A, Choi JY, van Wijnen AJ, Stein JL, Lian JB, Stein GS. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci. 2001;114:3093–3102. doi: 10.1242/jcs.114.17.3093. [DOI] [PubMed] [Google Scholar]
- 27.Olson MO, Hingorani K, Szebeni A. Conventional and nonconventional roles of the nucleolus. Internatl Rev Cytol. 2002;219:199–266. doi: 10.1016/S0074-7696(02)19014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma H, Siegel AJ, Berezney R. Association of chromosome territories with the nuclear matrix. Disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J Cell Biol. 1999;146:531–542. doi: 10.1083/jcb.146.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 30.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verschure PJ, van Der Kraan I, Manders EM, van Driel R. Spatial relationship between transcription sites and chromosome territories. J Cell Biol. 1999;147:13–24. doi: 10.1083/jcb.147.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 33.Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner S, Chiosea S, Nickerson JA. The spatial targeting and nuclear matrix binding domains of SRm160. Proc Natl Acad Sci U S A. 2003;100:3269–3274. doi: 10.1073/pnas.0438055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lian JB, Stein GS, Stein JL, van Wijnen AJ. Regulated expression of the bone-specific osteocalcin gene by vitamins and hormones. In: Litwack G, editor. Vitamins and Hormones. Academic Press; San Diego: 1998. pp. 443–509. [DOI] [PubMed] [Google Scholar]
- 36.Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- 37.Owen TA, Aronow M, Shalhoub V, Barone LM, Wilming L, Tassinari MS, Kennedy MB, Pockwinse S, Lian JB, Stein GS. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J Cell Physiol. 1990;143:420–430. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 38.Montecino M, Lian J, Stein G, Stein J. Changes in chromatin structure support constitutive and developmentally regulated transcription of the bone-specific osteocalcin gene in osteoblastic cells. Biochemistry. 1996;35:5093–5102. doi: 10.1021/bi952489s. [DOI] [PubMed] [Google Scholar]
- 39.Javed A, Gutierrez S, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D responsive transcription and contribute to chromatin organization. Mol Cell Biol. 1999;19:7491–7500. doi: 10.1128/mcb.19.11.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montecino M, Frenkel B, van Wijnen AJ, Lian JB, Stein GS, Stein JL. Chromatin hyperacetylation abrogates vitamin D-mediated transcriptional upregulation of the tissue-specific osteocalcin gene in vivo. Biochemistry. 1999;38:1338–1345. doi: 10.1021/bi982171a. [DOI] [PubMed] [Google Scholar]
- 41.Paredes R, Gutierrez J, Gutierrez S, Allison L, Puchi M, Imschenetzky M, van Wijnen A, Lian J, Stein G, Stein J, Montecino M. Interaction of the 1alpha,25-dihydroxyvitamin D3 receptor at the distal promoter region of the bone-specific osteocalcin gene requires nucleosomal remodelling. Biochem J. 2002;363:667–676. doi: 10.1042/0264-6021:3630667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paredes R, Arriagada G, Cruzat F, Villagra A, Olate J, Zaidi K, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. The bone-specific transcription factor RUNX2 interacts with the 1α,25-dihydroxyvitamin D3 receptor to up-regulate rat osteocalcin gene expression in osteoblastic cells. Mol Cell Biol. 2004;24:8847–8861. doi: 10.1128/MCB.24.20.8847-8861.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nangia AK, Butcher JL, Konety BR, Vietmeier BN, Getzenberg RH. Association of vitamin D receptors with the nuclear matrix of human and rat genitourinary tissues. J Steroid Biochem Mol Biol. 1998;66:241–246. doi: 10.1016/s0960-0760(98)00039-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhang C, Dowd DR, Staal A, Gu C, Lian JB, van Wijnen AJ, Stein GS, MacDonald PN. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J Biol Chem. 2003;278:35325–35336. doi: 10.1074/jbc.M305191200. [DOI] [PubMed] [Google Scholar]
- 45.Sierra J, Villagra A, Paredes R, Cruzat F, Gutierrez S, Javed A, Arriagada G, Olate J, Imschenetzky M, van Wijnen AJ, Lian JB, Stein GS, Stein JL. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol Cell Biol. 2003;23:3339–3351. doi: 10.1128/MCB.23.9.3339-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen J, Montecino MA, Lian JB, Stein GS, van Wijnen AJ, Stein JL. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D dependent, bone tissue-specific transcription. J Biol Chem. 2002;277:20284–20292. doi: 10.1074/jbc.M112440200. [DOI] [PubMed] [Google Scholar]
- 47.Carvallo L, Henriquez B, Olate J, van Wijnen A, Lian JB, Stein GS, Onate S, Stein JL, Montecino M. The 1alpha,25-dihydroxy vitamin D3 receptor preferentially recruits the coactivator SRC-1 during up-regulation of the osteocalcin gene. J Steroid Biochem Mol Biol. 2006 doi: 10.1016/j.jsbmb.2006.12.022. Ref Type: In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barletta F, Dhawan P, Christakos S. Integration of hormone signaling in the regulation of human 25(OH)D3 24-hydroxylase transcription. Am J Physiol Endocrinol Metab. 2004;286:E598–E608. doi: 10.1152/ajpendo.00214.2003. [DOI] [PubMed] [Google Scholar]
- 49.Dhawan P, Peng X, Sutton AL, MacDonald PN, Croniger CM, Trautwein C, Centrella M, McCarthy TL, Christakos S. Functional cooperation between CCAAT/enhancer-binding proteins and the vitamin D receptor in regulation of 25-hydroxyvitamin D3 24-hydroxylase. Mol Cell Biol. 2005;25:472–487. doi: 10.1128/MCB.25.1.472-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaisanen S, Dunlop TW, Sinkkonen L, Frank C, Carlberg C. Spatio-temporal activation of chromatin on the human CYP24 gene promoter in the presence of 1alpha,25-Dihydroxyvitamin D3. J Mol Biol. 2005;350:65–77. doi: 10.1016/j.jmb.2005.04.057. [DOI] [PubMed] [Google Scholar]
- 51.Staal A, van Wijnen AJ, Birkenhäger JC, Pols HAP, Prahl J, DeLuca H, Gaub MP, Lian JB, Stein GS, van Leeuwen JPTM, Stein JL. Distinct conformations of VDR/RXRα heterodimers are specified by dinucleotide differences in the vitamin D responsive elements of the osteocalcin and osteopontin genes. Mol Endocrinol. 1996;10:1444–1456. doi: 10.1210/mend.10.11.8923469. [DOI] [PubMed] [Google Scholar]
- 52.Shen Q, Christakos S. The vitamin D receptor, Runx2, and the Notch signaling pathway cooperate in the transcriptional regulation of osteopontin. J Biol Chem. 2005;280:40589–40598. doi: 10.1074/jbc.M504166200. [DOI] [PubMed] [Google Scholar]
- 53.Stein GS, Lian JB, Montecino M, van Wijnen AJ, Stein JL, Javed A, Zaidi K. Involvement of nuclear architecture in regulating gene expression in bone cells. In: Bilezikian JP, Raisz LG, Rodan GA, editors. Principles of Bone Biology. Academic Press; San Diego: 2002. pp. 169–188. [Google Scholar]
- 54.Schinke T, Karsenty G. Transcriptional control of osteoblast differentiation and function. In: Bilezikian JP, Raisz LG, Rodan GA, editors. Principles of Bone Biology. Academic Press; San Diego: 2002. pp. 83–91. [Google Scholar]


