Abstract
Structural analysis of the Escherichia coli response regulator transcription factor PhoB indicates that the protein dimerizes in two different orientations, both of which are mediated by the receiver domain. The two dimers exhibit two-fold rotational symmetry: one involves the α4-β5-α5 surface and the other involves the α1/α5 surface. The α4-β5-α5 dimer is observed when the protein is crystallized in the presence of the phosphoryl-analog BeF3− while the α1/α5 dimer is observed in its absence. From these studies a model of the inactive and active states of PhoB has been proposed that involves the formation of two distinct dimers. In order to gain further insight into the roles of these dimers we have engineered a series of mutations in PhoB intended to selectively perturb each of them. Our results indicate that perturbations to the α4-β5-α5 surface disrupt phosphorylation-dependent dimerization and DNA binding as well as PhoB mediated transcriptional activation of phoA while perturbations to the α1/α5 surface do not. Furthermore, experiments with a GCN4 leucine zipper/PhoB chimera protein indicate that PhoB is activated through an intermolecular mechanism. Together these results support a model of activation of PhoB in which phosphorylation promotes dimerization via the α4-β5-α5 face which in turn enhances DNA binding and thus the ability of PhoB to regulate transcription.
Keywords: two-component signal transduction, response regulator, alternate dimers, activation, conformational equilibrium
Introduction
Bacteria are routinely challenged by rapidly changing environments. The ability to effectively monitor their surroundings and alter their physiology in response to environmental changes is critical to competitiveness and survival. One strategy that bacteria employ to accomplish this adaptation is regulation via two-component signal transduction systems.1–3 Two-component systems (TCSs) abound in prokaryotes, while they are only minimally present in most eukaryotes. In fact, many eukaryotes, including all animals, have no TCS proteins encoded within their genomes. TCSs regulate a wide variety of physiological functions including motility, metabolic function, quorum sensing, and in many important human pathogens, virulence.4–7 The absence of these systems in humans, and their importance in many aspects of bacterial physiology, make TCSs attractive targets for development of novel antimicrobial therapeutics.8–12
The typical TCS consists of two conserved proteins, a histidine protein kinase (HK) and a response regulator (RR) that function together in a phosphotransfer pathway.1,3,13 The HK monitors the environment for specific cues and regulates the phosphorylation level of the RR in a stimulus-dependent manner. RRs are typically multi-domain proteins consisting of a conserved receiver (REC) domain and a variable effector domain.14 The role of the REC domain is to regulate the activity of the associated effector domain in a phosphorylation-dependent manner and the role of the effector domain is to mediate the response.
REC domains are highly versatile signaling modules that are capable of regulating the activities of a diverse array of effector domains.14 Although a variety of different types of effector domains have been identified in RRs, the most common type is a DNA-binding effector domain.14,15 RRs with DNA-binding effector domains can be sub-classified based on the specific fold of the DNA-binding domain. The most populated of these subfamilies is the OmpR/PhoB subfamily,14 named for its best known members and characterized by a winged helix-turn-helix DNA-binding domain.16–18
As is true for all RRs, the REC domain of OmpR/PhoB RRs regulates the activity of the protein by mediating distinct inter- and/or intramolecular interactions in the inactive and active states.19–23 Activation of RR transcription factors typically involves dimerization or higher order oligomerization.3,14,20,23–30 In several RRs the REC domain has been shown to mediate the formation of more than one type of dimer. For example, the REC domain of NtrC1, a transcription factor from Aquifex aeolicus, mediates the formation of two types of symmetric dimers. Each dimer differentially influences higher-order oligomerization of the molecule, an essential component of transcriptional activation. Domain arrangements in the inactive state dimer prevent oligomerization via the AAA+ ATPase domain while formation of the active state dimer relieves this inhibition.20 PleD, a diguanylate cyclase from Caulobacter crescentus, also forms distinct dimers in active and inactive states. Catalysis of cyclic di-GMP synthesis occurs only when the active sites of two PleD protomers are positioned in a specific orientation relative to each other, an orientation that is dependent on how the REC domains dimerize.30 The REC domain of Escherichia coli PhoB also forms two types of dimers.23,31
PhoB is the central regulator in the phosphate assimilation pathway in E. coli. Together with the HK PhoR, PhoB regulates expression of the Pho regulon, a set of ~40 genes that function in either the uptake or metabolism of a variety of different sources of phosphate.32–35 Under conditions of phosphate limitation, PhoB is phosphorylated; this enhances its affinity for DNA and allows it to regulate transcription of the Pho regulon. Despite extensive previous characterization of PhoB and structural information for both isolated REC and DNA-binding domains in various states,23,31,36–39 many fundamental questions regarding the molecular mechanisms involved in activation remain unanswered.
Biochemical studies have established a correlation between activation and dimerization,24,25 however, the specific roles of the two dimers are not clear. Although the intact form of PhoB has been reticent to crystallization, structural information is available for the isolated REC domain of PhoB in both the presence and absence of the non-covalent phosphoryl analog BeF3−.23,31 In the presence of BeF3− PhoB dimerizes with two-fold symmetry via the α4-β5-α5 face, while in its absence, a symmetric dimer mediated by the α1/α5 face is observed. In these two dimers, Thr83 and Tyr102, two residues that undergo a conserved repositioning upon activation,3,40 are in rotameric conformations associated with the active state in the α4-β5-α5 dimer23 and in rotameric conformations associated with the inactive state in the α1/α5 dimer31. This, coupled with the fact that the α4-β5-α5 dimer forms only in the presence of the phosphoryl analog BeF3−,23 suggests that the α4-β5-α5 dimer is the active state dimer. However, structural studies of two constitutively active PhoB mutant proteins have been interpreted as evidence that the α1/α5 dimer is the active state dimer.39
To ascribe activities to the two dimers and to gain further understanding of the role of dimerization in activation of PhoB, we have characterized proteins with single- and double-site substitutions in several of the residues that stabilize the two different dimers. Perturbation of the α4-β5-α5 surface disrupts phosphorylation-dependent DNA binding and the ability of PhoB to activate transcription, while perturbation of the α1/α5 surface does not. In addition, experiments carried out with a chimeric form of PhoB, in which the REC domain was replaced by a leucine zipper dimerization domain, suggest that dimerization alone is sufficient to activate the protein for transcriptional regulation. These results are consistent with the model of the active state proposed from structural analyses22,23 and suggest that dimerization is a critical feature for activation of PhoB and presumably for all members of the OmpR/PhoB subfamily.
Results
Rationale for mutagenesis
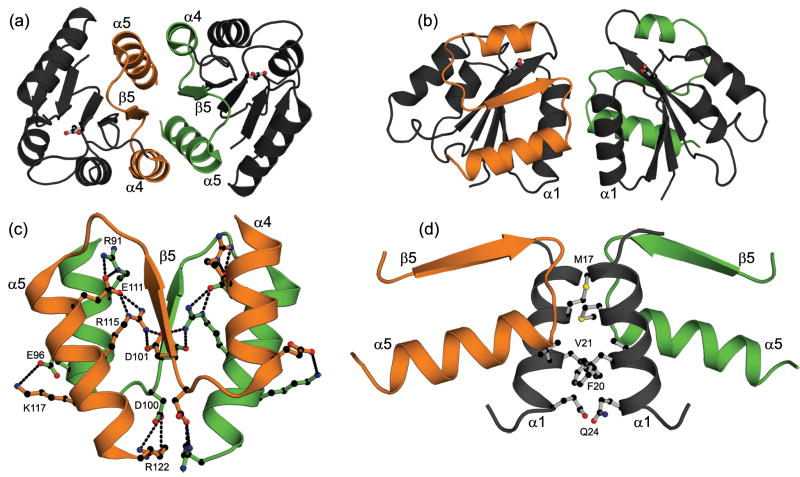
Specific mutations were made to PhoB with the goal of separately disrupting either the α4-β5-α5 dimer (Fig. 1a) or the α1/α5 dimer (Fig. 1b). The α4-β5-α5 dimer of PhoB is stabilized by a few hydrophobic interactions and an extensive network of salt bridges (Fig. 1c). Residues involved in these interactions are highly and exclusively conserved within the OmpR/PhoB subfamily28 and similar interfaces are observed in the structures of all other OmpR/PhoB family members crystallized in an active conformation.23,28,29,41,42 The α4-β5-α5 interface of PhoB contains four pairs of intermolecular salt bridges: R91…E111, D101…R115, E96…K117 and D100…R122 (Fig. 1c). The D101…R115 salt bridge was targeted to selectively disrupt the α4-β5-α5 dimer because it lies at the center of the interface (Fig. 1c). We reasoned that D101 and R115 contribute significantly to stabilization of the dimer because these residues are buried and do not compete with solvent molecules for hydrogen bonds as do some of the peripheral salt bridges (e.g. E96…K117) and R115 also participates in intramolecular ionic interactions with E111 and D101. D101 and R115 were separately mutated to the opposing residue of the salt bridge (PhoBD101R and PhoBR115D). In the context of the network, these single-residue mutations would result in repulsive charge-charge interactions that could potentially destabilize the dimer.
Fig. 1.
The two dimers formed by the REC domain of PhoB. A ribbon diagram of the α4-β5-α5 dimer (a) and the α1/α5 dimer (b) showing the different orientations of the two protomers within each dimer. The site of phosphorylation, D53, is shown in ball-and-stick representation. A detailed view of the α4-β5-α5 dimer interface (c) and the α1/α5 dimer interface (d). Several residues that stabilize the dimers are shown in ball-and stick representation. In panel (c) the intra- and intermolecular salt bridges are depicted as dashed lines and residues within one salt bridge of each pair are labeled. Throughout this figure the α4-β5-α5 face of one protomer is colored in green while the α4-β5-α5 face of the opposing protomer is colored in orange. Carbon atoms are colored black, oxygen atoms are colored red and nitrogen atoms are colored blue. Images in this figure were generated using PyMol (http://pymol.org/) and the coordinates for the α4-β5-α5 and α1/α5 dimers correspond to PDB codes 1ZES and 1B00, respectively.
In addition to engineering the two single-residue substitutions, we took advantage of the complementary electrostatics of the salt bridge and created a double substitution, PhoBD101R/R115D that would retain the potential to form favorable charge-charge interactions at the dimer interface. However, D101 and R115 participate in additional hydrophobic and ionic interactions that would not be recapitulated in the reversed pair (Fig. 1c); thus merely switching two residues within the ionic pair would not be expected to provide stabilization comparable to that of the wild-type protein.
The α1/α5 dimer of PhoB is mediated by residues from α1, the loop connecting β5 and α5, and the N terminus of α5 (Fig. 1d).23,31 Two residues, M17 and F20, comprise 30% of the buried solvent accessible surface area of the interface. In an attempt to destabilize the α1/α5 dimer, F20 was chosen as a mutagenesis target because it not only stabilizes the dimer through hydrophobic interactions with V21 and the aliphatic portion of the Q24 side chain, but the aromatic rings of the F20 residues of the two protomers pack together at the dimer interface (Fig. 1d). We reasoned that we could effectively disrupt the interface by replacing F20 with an aspartate (PhoBF20D), eliminating the hydrophobic interactions and introducing electrostatic repulsion.
Phosphotransfer activity of mutant PhoB proteins
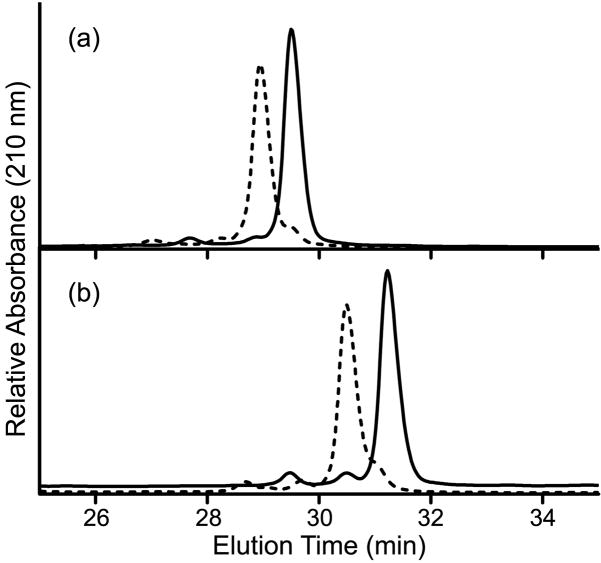
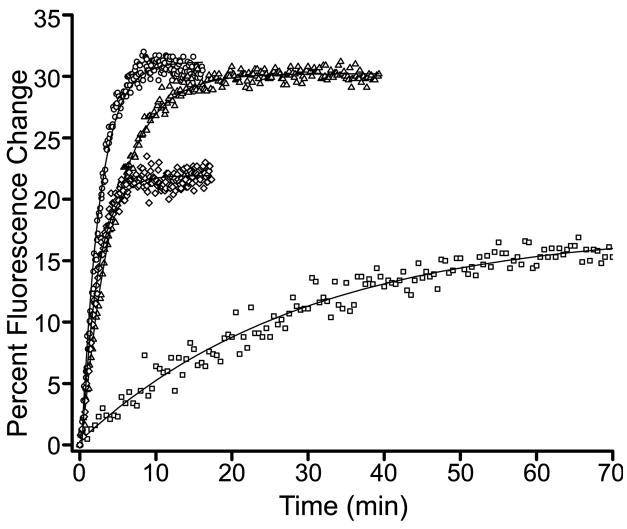
The activity of PhoB as a transcription factor is dependent on its phosphorylation state. Thus, it was important to determine whether the mutant proteins exhibited normal phosphorylation prior to evaluating their DNA-binding and transcriptional regulation activities. Phosphotransfer activities of purified PhoB proteins (wild-type PhoB, PhoBR115D, phoBD101R/R115D and PhoBF20D) were assayed in vitro using phosphoramidate as the phosphodonor.43 PhoBD101R was excluded from these assays as it formed inclusion bodies under all expression conditions examined and could not be obtained as a native purified protein. Phosphorylation was monitored by mobility shifts during reverse-phase HPLC (Fig. 2) and by real-time fluorescence spectroscopy (Fig. 3), exploiting the phosphorylation-induced perturbation in intrinsic tryptophan fluorescence.25 Upon incubation of wild-type PhoB, PhoBD101R/R115D and PhoBF20D with phosphoramidate, >90% of the protein became phosphorylated, typically within 45 min (Fig. 2 and Table 1). Furthermore, these three proteins exhibited similar phosphorylation rates (Fig. 3 and Table 1). In contrast, PhoBR115D achieved a steady-state phosphorylation level of only 60% (Table 1), and required ~90 min to attain that level. The incomplete phosphorylation of PhoBR115D is not the result of a significant percentage of the protein existing in an unfolded or otherwise non-functional form, since a greater percentage of PhoBR115D is phosphorylated in the presence of a higher concentration of phosphoramidate (data not shown). Rather, the lower steady-state level of phosphorylation observed for PhoBR115D can be explained by a slower rate of phosphorylation.
Fig. 2. Representative reverse-phase HPLC profiles of unphosphorylated and phosphorylated PhoB.
Untreated (solid line) and phosphoramidate-treated (dashed line) wild-type PhoB (a) and PhoBD101R/R115D (b) were analyzed on a reverse-phase HPLC column as described in Materials and Methods. The steady-state phosphorylation levels attained for wild-type PhoB, PhoBD101R/R115D, PhoBR115D and PhoBF20D are listed in Table 1.
Fig. 3. Real-time intrinsic fluorescence analysis of PhoB.
The intrinsic fluorescence changes that occur as a function of phosphorylation were monitored for wild-type PhoB (circles), PhoBD101R/R115D (triangles), PhoBR115D (squares) and PhoBF20D (diamonds). The observed phosphorylation rates (kobs) are listed in Table 1.
Table 1.
Autophosphorylation of PhoB proteins using phosphoramidate as donor
| Protein | % Phosphorylation (steady-state)a | Phosphorylation Rate, kobs (s−1)b |
|---|---|---|
| Wild-type PhoB | 93 | 7.0 ±0.1 × 10−3 |
| PhoBR115D | 60 | 0.6 ±0.1 × 10−3 |
| PhoBD101R/R115D) | 92 | 3.4 ± 0.5 × 10−3 |
| PhoBF20D | 93 | 7.9 ± 0.2 × 10−3c |
| PhoBR91E/E111R | 100 | 10.1 ±0.2 × 10−3c |
The steady-state phosphorylation level was determined by HPLC analysis.
The rate of phosphorylation was measured by monitoring intrinsic protein fluorescence changes.
The error refers to the goodness of the fit for only one replicate, for all other kobs values the error represents one standard deviation from a minimum of two replicates.
Characterization of PhoB dimerization
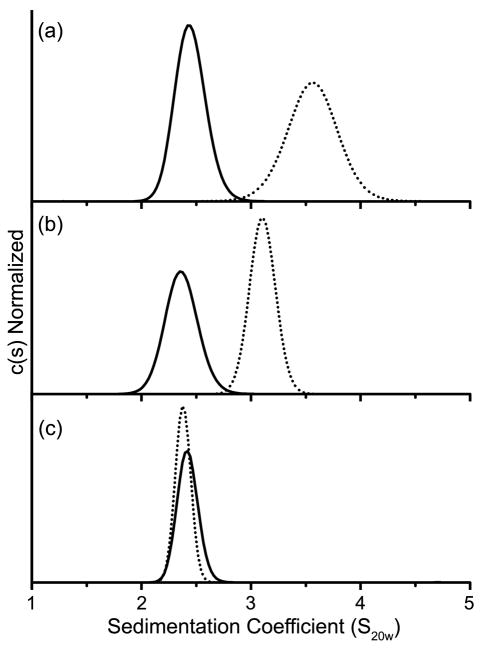
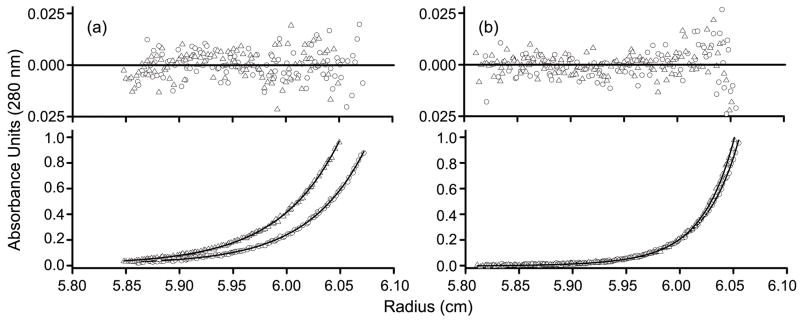
Analytical ultracentrifugation was employed to determine the effects on dimerization of mutations within the α4-β5-α5 or α1/α5 interfaces. Sedimentation velocity (SV) and sedimentation equilibrium (SE) experiments were initially performed on both the unphosphorylated and phosphorylated forms of wild-type PhoB. SV data were analyzed by generating a continuous sedimentation [c(s)] distribution (Fig. 4), which is a depiction of the weight-average sedimentation coefficient (Swa) of all the species present.44 SE data were analyzed by fitting six SE profiles (three concentrations at two speeds) to either a single-species model or a monomer-dimer model. When the unphosphorylated form of wild-type PhoB was analyzed by SV at a loading concentration of 13 μM, a single peak was observed with an Swa characteristic of a monomer (~2.6 S) (Fig. 4a). Although only a single peak was observed, PhoB exhibited an increase in Swa as a function of increased loading concentration (data not shown), indicating that the monomer form of unphosphorylated PhoB is in equilibrium with a higher order species. In order to quantitatively characterize this equilibrium, wild-type PhoB was analyzed by SE at concentrations ranging from 5–20 μM. A monomer-dimer model with a Kd of 378 ± 102 μM fits the SE profiles with the lowest χ2 (Fig. 5a and Table 2). SE analysis of phosphorylated wild-type PhoB also indicated a monomer-dimer equilibrium (Fig. 5b), however, the dimerization affinity of the phosphorylated form is ~100 fold greater than that of the unphosphorylated form (Table 2).
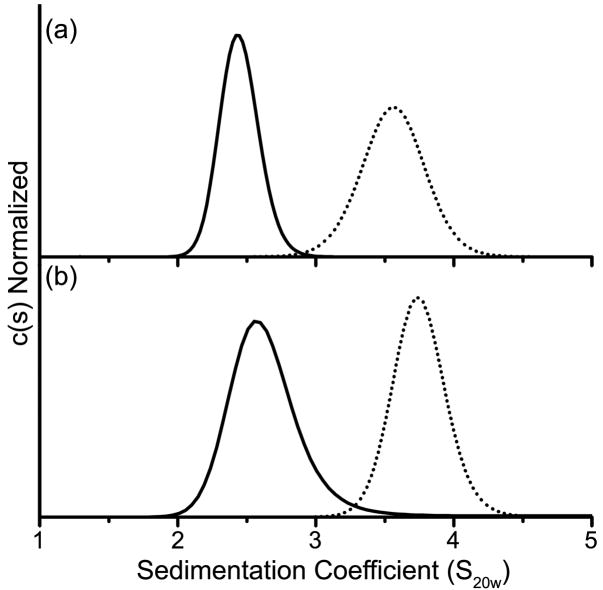
Fig. 4. Continuous sedimentation [c(s)] distributions of phosphorylated and unphosphorylated PhoB.
Wild-type PhoB (a), PhoBD101R/R115D (b) and PhoBR115D (c) were analyzed in both their unphosphorylated (solid line) and phosphorylated (dotted line) forms as described in Materials and Methods.
Fig. 5. Representative sedimentation equilibrium profiles of wild-type PhoB and PhoBF20D.
Wild-type PhoB (circles) and PhoBF20D (triangles) were analyzed in both their unphosphorylated (a) and phosphorylated (b) forms. Profiles are shown for the 20 μM samples that were spun at 30k RPM with the corresponding residuals of the fits shown above the profiles. For unphosphorylated and phosphorylated wild-type PhoB, as well as phosphorylated PhoBF20D, the monomer-dimer fit is shown, while for unphosphorylated PhoBF20D the monomer fit is shown.
Table 2.
Sedimentation equilibrium analysis of PhoB proteins
| Protein | Unphosphorylated | Phosphorylated | ||
|---|---|---|---|---|
| Modela | Kd or MW | Modela | Kd or MW | |
| Wild-type PhoB | M-D | 378 ± 102 μM | M-D | 5.1 ± 1.1 μM |
| PhoBF20D | M | 26521 ± 379 Da | M-D | 2.8 ± 0.8 μM |
| PhoBZ | M-D | 0.91 ± 0.78 μMb | N/Ac | N/Ac |
SE profiles were fit to a monomer (M) model and a monomer-dimer (MD) model; the model that fit each data set with the lowest χ2 is listed above. For the monomer model the molecular weight (MW) was treated as a floating parameter and for the M-D model the dissociation constant (Kd) was treated as a floating parameter.
The large errors associated with this measurement likely reflect the high dimerization affinity of PhoBZ. A lower concentration of protein, which is not feasible with the absorbance detection system, would be required to obtain a more definitive value.
Not applicable.
The two proteins with mutations targeted to the α4-β5-α5 interface, PhoBR115D and PhoBD101R/R115D, were characterized by SV (Fig. 4). Wild-type PhoB and PhoBD101R/R115D were analyzed at a concentration of 13 μM while PhoBR115D was analyzed at a concentration of 25 μM. A higher molar concentration was used for PhoBR115D because only 60% of the protein was phosphorylated compared to wild-type PhoB and PhoBD101R/R115D, which attained a phosphorylation level of >90% (Table 1). At these concentrations the initial molar amount of phosphorylated protein is approximately equivalent for all three proteins. The unphosphorylated forms of wild-type PhoB, PhoBD101R/R115D and PhoBR115D exhibit similar c(s) profiles that are all characteristic of a monomer (Fig. 4). Given the protein loading concentrations that were used for the SV analyses (5–25 μM) and the Kd of unphosphorylated wild-type PhoB (378 ± 102 μM), it is not surprising that the predominant species present during the SV run is a monomer. When the proteins were incubated with phosphoramidate there was a shift to a larger S value for both wild-type PhoB and PhoBD101R/R115D. Wild-type PhoB exhibited the largest shift, suggesting that within the population there is a higher percentage of dimer than exists for phoBD101R/R115D (Fig. 4). The c(s) profiles for unphosphorylated and phosphorylated PhoBR115D are similar, which indicates that PhoBR115D does not dimerize upon phosphorylation, at least not to the same extent as wild-type PhoB and PhoBD101R/R115D. It is important to note that over the time course of the SV run (~9 h) the steady-state level of phosphorylated PhoBD101R/R115D and PhoBR115D decreased to ~40% while the level of phosphorylated wild-type PhoB remained close to 90% (HPLC unpublished results). However, the absence of a shift for PhoBR115D is not likely due to a decreased level of phosphorylation, since a shift in Swa is observed for PhoBD101R/R115D, which decreases to a similar level of phosphorylation.
SE was employed to determine the effect on dimerization of the F20D substitution. In contrast to wild-type PhoB, PhoBF20D did not dimerize in the unphosphorylated state. SE profiles for unphosphorylated PhoBF20D were fit to both a monomer model and a monomer-dimer model. The monomer model yielded a molecular weight of 26521 ± 379 Da, which is similar to the theoretical molecular weight of a monomer (26401 Da). Furthermore, there was not a statistically significant improvement in the fit when a monomer-dimer model was used. Although the aspartic acid substitution affects dimerization when the protein is unphosphorylated, the Kd for the phosphorylated form (2.8 ± 0.8 μM) is similar to that of wild-type PhoB (Table 2).
DNA-binding analysis
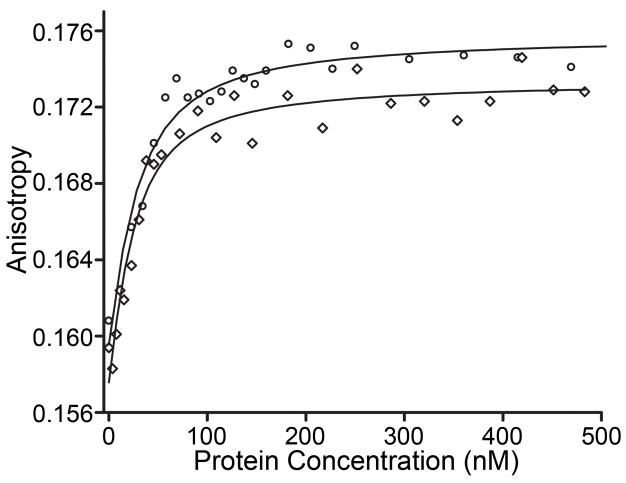
Previous studies have shown that phosphorylation enhances the affinity of PhoB for DNA by ~10 fold.25 Our fluorescence anisotropy results are in agreement with these early studies. Phosphorylation enhances the affinity of PhoB for a synthetic oligonucleotide containing the pho box portion of the phoB promoter (Fig. S1) by ~15 fold. The Kd of unphosphorylated PhoB is 145 ± 30 nM compared to 9.7 ± 2.2 nM when it is phosphorylated (Fig. 6 and Table 3). Similarly, phosphorylation significantly enhances the DNA-binding affinity of both PhoBF20D and PhoBD101R/R115D. Phosphorylation of PhoBF20D increases its affinity for DNA by ~50 fold. PhoBD101R/R115D binds non-specifically when unphosphorylated and specifically with a Kd of 23.1 ± 6.3 nM when it is phosphorylated (Table 3). The increased DNA-binding affinity presumably results from dimerization upon phosphorylation (Table 2).25 In contrast, PhoBR115D does not display phosphorylation-enhanced DNA binding (Table 3). Thus under conditions that favor phosphorylation, perturbations to the α4-β5-α5 surface, and not the α1/α5 surface, decrease both dimerization and DNA binding. Furthermore, compensatory double residue mutations (PhoBD101R/R115D) are less deleterious to DNA binding than a single residue substitution (PhoBR115D), suggesting that a salt bridge between residues 101 and 115 on the α4-β5-α5 face promotes dimerization and DNA binding.
Fig. 6. Representative fluorescence anisotropy DNA-binding curves.
Binding isotherms for the phosphorylated forms of wild-type PhoB (circles) and PhoBF20D (diamonds) are shown to illustrate typical fluorescence anisotropy binding curves. A synthetic oligonucleotide (Fig. S1) that contains a portion of the phoB promoter was used in these experiments. Dissociation constants for all the proteins analyzed are listed in Table 3.
Table 3.
DNA binding activities of PhoB proteins
| Protein | Kd(nM), Unphosphorylateda | Ka (nM), Phosphorylateda |
|---|---|---|
| Wild-type PhoB | 145 ± 30 | 9.7 ± 2.2 |
| PhoBD101R/R115D | NSb | 23.1 ± 6.3 |
| PhoBR115D | 256 ± 41 | 183 ± 18 |
| PhoBF20D | 457 ± 50 | 8.3 ± 1.4 |
| PhoBR91E/E111R | 86.2 ± 2.6 | 2.9 ± 0.3 |
The error represents one standard deviation from a minimum of three replicates.
Non-specific binding.
Transcription regulation activity of PhoB
The gene for alkaline phosphatase (phoA) is one of the genes that comprise the Pho regulon.32,35 In order to assess the transcriptional regulation activity of the various mutant PhoB proteins, alkaline phosphatase levels (AP) were measured in cells containing a chromosomal deletion of phoB (Genobase ID: JW038945). The various PhoB proteins were expressed from plasmids under control of the lac promoter. IPTG was used to control PhoB protein levels such that they were within two fold of the level observed in wild-type cells under low phosphate conditions as assessed by quantitative Western blot analysis. Our analyses of endogenously expressed wild-type PhoB indicated the intracellular molar concentrations of PhoB under non-inducing and inducing conditions to be ~1 and 10μM, respectively (Table S1 and Fig. S2).
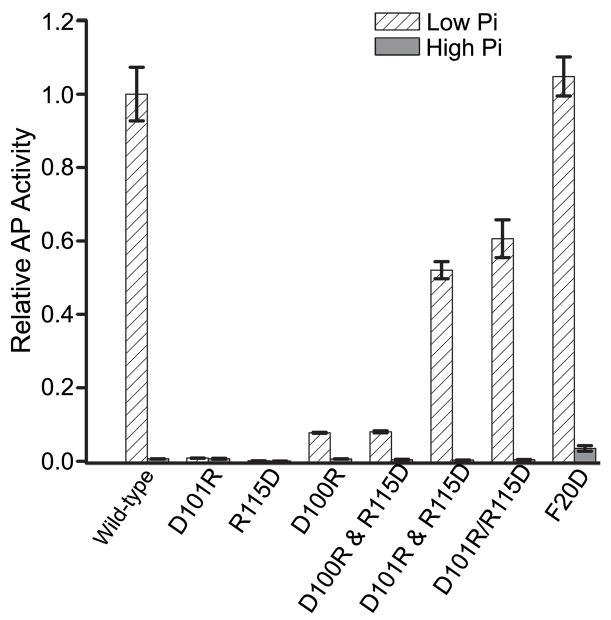
As has been observed previously, wild-type PhoB exhibited phosphate-dependent regulation of phoA (Fig. 7). The transcriptional activity of PhoBF20D, which targets the al/a5 surface, is similar to that of wild-type PhoB (Fig. 7). In contrast, PhoBD101R and PhoBR115D, both of which target the α4-β5-α5 surface, do not activate transcription of phoA while PhoBD101R/R115D retains the ability to activate phoA transcription in a phosphate-dependent manner (Fig. 7). Thus it appears that the repulsive effects of the single-residue substitutions are more deleterious to activation of PhoB than the compensatory double substitutions. Furthermore, when PhoBR115D and PhoBD101R were expressed in the same cell from two different plasmids, the AP levels were the same as for PhoBD101R/R115D (Fig. 7). Co-expression of PhoBD100R and PhoBR115D, with mutations in residues that do not interact with each other in the interface, did not activate transcription of phoA. In addition to providing insight into the importance of the D101…R115 interaction, this result also indicates that PhoBR115D is properly folded in vivo.
Fig. 7. Transcriptional activation of phoA by PhoB mutants.
Cells expressing the various mutants of PhoB were grown under low phosphate (inducing) and high phosphate (non-inducing) conditions and analyzed for AP activity as described in Materials and Methods. The “&” symbol is used to identify those cells that express two mutants from different plasmids while a “/” symbol is used to designate the double-residue mutant. Each value represents a minimum of 3 replicates with error bars representing one standard deviation. The AP activity was normalized to the level attained by wild-type PhoB under low phosphate conditions.
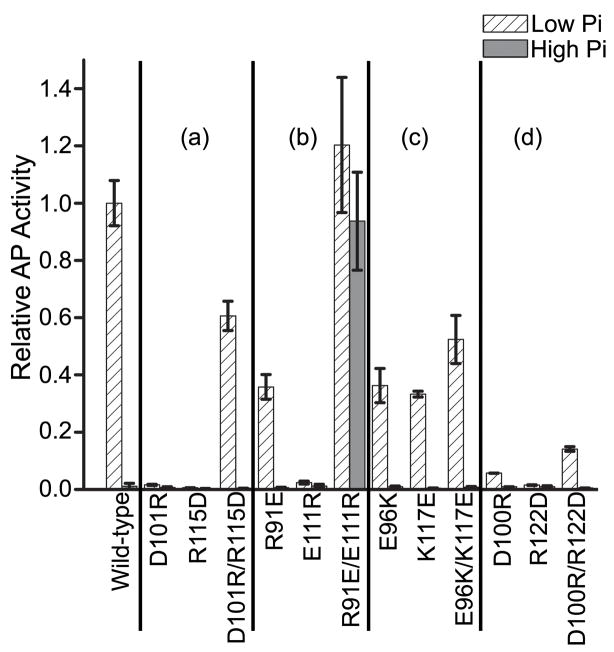
The D101…R115 salt bridge is only one of several interactions that stabilize the α4-β5-α5 dimer (Fig. 1c). In order to gain a more complete understanding of the role of other conserved residues in PhoB activation, two disruptive single-residue mutations and one potentially compensatory double-residue mutation pair were engineered in PhoB to target each of the other three intermolecular salt bridges (Fig. 1c). Like cells expressing PhoBD101R or PhoBR115D, cells expressing PhoBD100R, PhoBE111R or PhoBR122D exhibit little or no activity under any condition (Fig. 8). In contrast, the three mutant proteins, PhoBR91E, PhoBE96K and PhoBK117E, exhibit significant, although reduced, activity. Interestingly, PhoBE96K and PhoBK117E contain mutations that target the same salt bridge and show similar AP activities, whereas the two mutations that target the R91…E111 salt bridge have very different effects. Proteins containing compensatory double-residue mutations all retain the ability to activate phoA expression, although in most cases to a lesser extent than wild-type PhoB. In all cases, the compensatory double-residue PhoB mutant proteins are more active than proteins containing either of the single-residue mutations that correspond to the same salt bridge (Fig. 8).
Fig. 8. Transcriptional activation of phoA by the single- and double-residue mutants that target the α4-β5-α5 surface.
Cells expressing the various mutants of PhoB were grown under low phosphate (inducing) and high phosphate (non-inducing) conditions and analyzed for AP activity as described in Materials and Methods. The “/” symbol is used to designate the double-residue mutants. For clarification, the results are grouped based on the salt bridge that they target. Group (a): D101…R115, group (b): R91…E111, group (c): E96…K117 and group (d): D100…R122. Each value represents a minimum of 3 replicates with error bars representing one standard deviation. The AP activity was normalized to the level attained by wild-type PhoB under low phosphate conditions.
One doubly-substituted protein, PhoBR91E/E111R, was found to constitutively activate transcription of phoA (Fig. 8). The in vitro analysis performed on this protein indicates that it not only binds to DNA with greater affinity than wild-type PhoB (Table 3), but also has a greater rate of phosphorylation (Table 1). This suggests that these residue substitutions have altered the equilibrium between inactive and active states, driving the conformational equilibrium toward the active state.46 Given the strong correlation between dimerization and activation, analytical ultracentrifugation was employed to determine if PhoBR91E/E111R is a constitutive dimer (Fig. 9). In the unphosphorylated state, the c(s) peak of PhoBR91E/E111R is broader and shifted to a slightly larger S value (Swa = 2.8) relative to wild-type PhoB, consistent with monomer–dimer equilibria and a larger population distribution of dimer for PhoBR91E/E111R than for wild-type PhoB (Swa = 2.6), which exists primarily as a monomer. Phosphorylation causes shifts of both proteins to larger sedimenting species, with the peak for PhoBR91E/E111R being narrower and shifted to a slightly larger S value (Swa = 4.0) relative to wild-type PhoB, consistent with population distributions of primarily dimer for PhoBR91E/E111R and a slightly larger percentage of monomer for wild-type PhoB (Swa = 3.8). Phosphorylation of PhoBR91E/E111R causes a shift to a larger sedimenting species that is slightly greater than wild-type PhoB. While this analysis is not rigorously quantitative, the data suggest that PhoBR91E/E111R dimerizes with a greater affinity than wild-type PhoB in both unphosphorylated and phosphorylated states, but that it is not a constitutive dimer.
Fig. 9. Sedimentation velocity analysis of PhoBR91E/E111R.
Wild-type PhoB (a) and PhoBR91E/E111R (b) were analyzed in both their unphosphorylated (solid line) and phosphorylated (dotted line) forms. The protein phosphorylation procedure as well as the details of the c(s) analysis are described in Materials and Methods.
The dimerization propensities of the mutant proteins indicate that phosphorylation-dependent dimerization of PhoB is driven by the REC domain, as has been previously demonstrated.24 This result and the postulated lack of interactions between the REC domain and the DNA-binding domain in the active state23,28,29 raise the possibility that the primary role of the REC domain in the activation of PhoB is to function as a dimerization domain. This hypothesis was tested by creating a chimera of PhoB (PhoBZ) in which the REC domain was replaced with a well characterized dimerization domain (Fig. S3).
The GCN4 leucine zipper motif was fused to the DNA-binding domain of PhoB (residues 128–229) via a 13-residue linker (ESLHPPMDEFRGS). The GCN4 leucine zipper forms a parallel coiled coil47 that would position the attached DNA-binding domains in close proximity, similar to their positions in intact active PhoB dimerized through the α4-β5-α5 face of the REC domain. The linker of PhoBZ was designed to be longer than the 5-residue linker present in PhoB in order to minimize constraints on the orientations of the DNA-binding domains, allowing for the potentially different positioning of points of attachment to the REC and leucine zipper dimerization domains. The oligomeric state of PhoBZ was analyzed by SE, yielding a Kd of 0.91 ± 0.78 μM, a dimerization affinity ~5-fold greater than phosphorylated wild-type PhoB (Table 2).
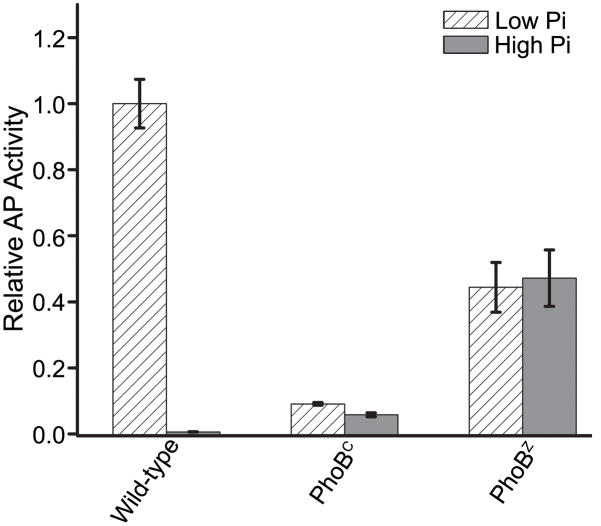
In contrast to intact PhoB, both PhoBZ and the isolated DNA-binding domain of PhoB (PhoBC) activate transcription of phoA in a phosphate-independent manner (Fig. 10). The constitutive activity is not surprising given the absence of the REC domain, the locus of phosphorylation-mediated dimerization.24,25,48 The activity exhibited by PhoBZ is ~50% of that observed for wild-type PhoB under low phosphate conditions, indicating that PhoBZ activates transcription of phoA less effectively than the phosphorylated intact protein. However, the activity of PhoBZ is ~5 fold greater than that observed for PhoBC (Fig. 10).
Fig. 10. Transcriptional activation of PhoBZ and PhoBC.
Cells expressing PhoBZ or PhoBC were grown under low phosphate (inducing) and high phosphate (non-inducing) conditions and analyzed for AP activity as described in Materials and Methods. Each value represents a minimum of 3 replicates with error bars representing one standard deviation. The AP activity was normalized to the level attained by wild-type PhoB under low phosphate conditions.
While the AP activity of PhoBC is significantly lower than that of PhoBZ, PhoBC is more active than wild-type PhoB under non-inducing conditions (Fig. 10). This observation was initially made over a decade ago and established the inhibitory role of the REC domain with regard to DNA binding and transcriptional activation.48 While qualitatively similar, our results differ quantitatively from those reported previously. Under non-inducing conditions, we observe that PhoBC has 5–8 fold greater activity than wild-type, less than the 15–20 fold difference observed previously.48
In order to determine whether the difference might be due to protein expression levels, wild-type PhoB and PhoBC were grown under high phosphate conditions at different IPTG concentrations, resulting in different expression levels. The activities of both wild-type PhoB and PhoBC increased with increasing expression levels, but to different extents (Fig. S4). Thus interestingly, the relative activities of wild-type PhoB and PhoBC varied as a function of expression level, with the greatest differences observed at the highest levels of expression. In this study, expression levels of mutant proteins were matched to the level of endogenously expressed wild-type PhoB under inducing conditions, in contrast to much higher levels of expression used in the previous study.48 The differences in the relative activities of PhoB and PhoBC observed in the different studies are likely a result of this concentration-dependent phenomenon. This phenomenon might be relevant to RRs other than PhoB and perhaps contributes to differences in the relative magnitude of inhibitory activity ascribed to REC domains of different RRs in previous studies. Our results suggest that for comparative purposes, in vivo activities should be determined at levels of proteins matched not only to each other but also to physiological levels.
Discussion
Amino acid substitutions to the α4-β5-α5 surface disrupt PhoB activation
To gain a more complete understanding of the roles of the two dimers in the activity of PhoB, we engineered and characterized PhoB proteins containing a set of amino acid substitutions intended to selectively perturb each of the two dimer interfaces. Our data indicate that inactive unphosphorylated PhoB exists primarily as a monomer in equilibrium with a small amount of α1/α5 dimer and that active phosphorylated PhoB exists primarily as an α4-β5-α5 dimer. Both in vitro and in vivo analyses indicate that substitutions within the α4-β5-α5 surface, and not the α1/α5 surface, perturb phosphorylation-dependent dimerization and DNA binding, as well as transcriptional activation of pho A.
These results support a model of the active state that evolved from structural studies of the isolated REC domain and DNA-binding domain of PhoB.23,37 In the active state, PhoB is presumed to exist as a dimer in which the REC domains associate with rotational symmetry via the α4-β5-α5 surface while the DNA-binding domains are tandemly oriented in order to interact with the direct repeat DNA sequences that comprise the PhoB recognition site.37,49 A similar active state has been postulated for all OmpR/PhoB transcription factors since residues that stabilize the α4-β5-α5 dimer are conserved within the OmpR/PhoB subfamily and this particular dimer has been observed in all the structures of activated OmpR/PhoB RR REC domains reported to date.23,28,29,41,42
While structure-driven site-directed mutagenesis has been, and continues to be, an important tool to probe protein-protein interactions, unintended consequences of mutations can complicate interpretation of such studies. When PhoP, an OmpR/PhoB RR from Bacillus subtilis, was crystallized, an asymmetric dimer mediated by the α4-β5-α5 and α2-α3-α4 surfaces was observed.50 Mutation of R113, a residue involved in stabilization of the dimer, prevented phosphorylation-dependent dimerization and DNA-binding activity, leading the authors to suggest that the asymmetric dimer observed in the crystal structure is the active form of the protein.51 However, R113 in PhoP corresponds to one of the conserved residues that stabilize the symmetric α4-β5-α5 dimer that has been observed in all characterized active OmpR/PhoB RR REC domains. In retrospect, it seems likely that the loss of activity resulting from substitution of R113 is due to disruption of the symmetric α4-β5-α5 active state dimer, rather than the intended disruption of the asymmetric dimer, which presumably corresponds to the inactive state.
Given that mutations designed to simply disrupt a dimer might yield ambiguous results, we took additional measures to minimize the likelihood of misinterpretation. First, the native folded states of PhoBR115D, PhoBD101R/R115D and PhoBF20D were verified by analysis of phosphotransfer activity. The low steady-state phosphorylation level of PhoBR115D was not a result of a significant percentage of the protein existing in an unfolded state, rather, this mutant autophosphorylates more slowly than the other proteins (see below). Second, compensatory mutations were engineered to probe the complementary nature of the salt bridge. PhoBD101R/R115D binds DNA and dimerizes in a phosphorylation-dependent manner while PhoBR115D does not. Furthermore, only when PhoBR115Dand PhoBD101R are co-expressed is phosphate-dependent activation of phoA restored. This suggests that the pairwise intermolecular contact between residues 101 and 115 is necessary for activation of the protein. The different activities of PhoBD101R/R115D and PhoBR115D, coupled with the fact that PhoBF20D does not dimerize in the unphosphorylated state but does exhibit phosphorylation-dependent dimerization and DNA binding, is strong evidence that activation of PhoB requires dimerization via the α4-β5-α5 surface.
Mutations at the α4-β5-α5 dimer interface have different impacts on transcriptional activation
In addition to targeting the D101…R115 salt bridge, mutations were made to the residues that form the other three salt bridges that stabilize the α4-β5-α5 dimer: R91…E111, E96…K117, D100…R122. In each case the transcriptional activity of the compensatory double-residue mutant protein was greater than that of either protein containing a single-residue substitution affecting the same salt bridge. These results are consistent with our model. However, the relative activity of these mutants varied significantly. Although the crystal structure cannot provide a rigorous quantitative assessment of the energetics of a given interaction, it can provide some insight into the causes of this variability.
Although a majority of the single-residue mutations that target the α4-β5-α5 surface result in proteins that are almost completely inactive, three proteins (PhoBR91E, PhoBE96K and PhoBK117E) exhibit significant activity. Interestingly, the residues that are mutated in these three proteins are all solvent exposed. Any interaction formed by solvent exposed residues will be weakened by competition with solvent molecules. Thus the solvent exposed residues might be expected to contribute less than the buried residues to stabilization of the dimer. An additional contribution to the variability likely stems from the complex network of ionic interactions. For example, D101 not only forms an intermolecular salt bridge with R115 but also forms intramolecular interactions with R115 and E111. In the case of PhoBD101R/R115D, the potential to form the intermolecular salt bridge might still exist but these substitutions will likely disrupt the intramolecular salt bridge between residues 115 and 111. Given the extensive network of interactions at the dimer interface, the broad range of activities observed is not surprising.
Mutations at the α4-β5-α5 alter the equilibrium between active and inactive states
RRs exist in equilibrium between inactive and active states with phosphorylation shifting this equilibrium towards the active state.52,53 Structural studies have shown that target binding also can influence the position of the equilibrium.54,55 In the case of the unphosphorylated E. coli RR CheY, binding to a peptide that corresponds to the FliM motor protein stabilizes a meta-active conformation.56 Furthermore, this interaction enhances the autophosphorylation rate of CheY.57 A similar effect is also observed with Salmonella enterica CheY when it binds a peptide that corresponds to the C-terminal helix of CheZ.55,57 In both cases it is presumed that target binding decreases the energy barrier of phosphorylation by stabilizing the active conformation.
Examples of stabilization of an active conformation can also be found within the OmpR/PhoB subfamily. In the crystal structures of ArcA, TorR and KdpE, the proteins dimerize symmetrically through the α4-β5-α5 face and are in the active conformation even in the absence of the phosphoryl analog BeF3−.28,29 In these cases the active conformation is likely stabilized through formation of the α4-β5-α5 dimer.
Mutations can alter the equilibrium between inactive and active conformations.54,58 An altered equilibrium can explain the behavior of the substituted PhoB proteins. PhoBR91E/E11R displayed constitutive activation of transcription. Additionally, PhoBR91E/E111R autophosphorylates more rapidly and exhibits a larger Swa compared to wild-type PhoB. Thus PhoBR91E/E111R has a greater propensity to dimerize, and this might stabilize the active conformation, increasing the rate of autophosphorylation. In contrast, PhoBR115D does not activate transcription, it autophosphorylates slower than wild-type PhoB and it is a monomer in both unphosphorylated and phosphorylated forms. PhoBR115D is not capable of dimerizing through the α4-β5-α5 surface, thus decreasing the subpopulation that exists in an active conformation and hence decreasing the rate of autophosphorylation.
Correlation of dimer structures with the active state
Several different crystal structures of the isolated REC domain of PhoB have been reported including both wild-type and mutant forms. In addition to apo wild-type PhoB, which crystallized in three different space groups,23,31,38 there are structures of wild-type PhoB bound to Mg+2 38 and Mg+2/BeF3−.23 Only in the PhoB-Mg+2/BeF3− structure is the α4-β5-α5 dimer observed; the α1/α5 dimers are observed in all other structures. Notably, the α1/α5 and α4-β5-α5 dimers can be crystallized under nearly identical conditions, with the only difference being the absence or presence of Mg+2/BeF3−, respectively. These structures suggest that the active state of PhoB corresponds to the α4-β5-α5 dimer. Our mutational analysis supports this hypothesis. However, two mutant proteins that are constitutively and strongly active (PhoBD53A/Y102C and PhoBD10A/D53E) both crystallize as α1/α5 dimers, leading to the proposal that the α1/α5 dimer is the active state.39
This apparent contradiction can be resolved by acknowledging that neither intracellular transcriptional activities nor conformations observed in crystal structures correlate linearly with the conformational distribution in a population of molecules. It is important to note that a constitutive phenotype does not require a protein to be locked in an active state. Specifically, in the case of PhoB, the level of active PhoB required to generate constitutive activity in a particular physiological assay has not been determined. Furthermore, because of complex selective forces during crystallization, the conformation that crystallizes need not reflect the dominant species present in solution. Indeed, constitutively active RR variants, E. coli CheYD13K and cheYD13K/Y106W, have been found to crystallize in inactive conformations.54,59
Biological roles of the α4-β5-α5 and α1/α5 dimers
The role of the phosphorylated REC domain of PhoB in activation of transcription was assessed by characterization of a chimeric protein in which the REC domain was replaced with a leucine-zipper dimerization domain. This chimera, PhoBZ, exhibited stimulus-independent activation of gene expression at a level ~half that of phosphorylated PhoB and ~5 fold greater than that of the isolated DNA-binding domain, PhoBC. These results indicate that the primary role of the phosphorylated REC domain is to promote dimerization of the DNA-binding domains. Furthermore, recapitulation of activity in the absence of the REC domain confirms an intermolecular mechanism of activation and provides strong evidence against previously postulated allosteric communication between the DNA-binding and phosphorylated REC domains in the active state.60,61
Although a leucine zipper can substitute for the phosphorylated REC domain to promote dimerization, PhoBZ is not as active as phosphorylated PhoB. Explanations for this include: 1) inactivity due to misfolding of PhoBZ; 2) weaker dimerization of the leucine zipper compared to phosphorylated REC domains; 3) suboptimal orientation of the DNA-binding domains in PhoBZ; and 4) inability of the leucine zipper to recapitulate possible interactions of the phosphorylated REC domain with polymerase or other components. Misfolding is an unlikely explanation for lower activity as PhoBZ behaved well when overexpressed and purified, eluting as a single peak during size exclusion chromatography and fitting a monomer-dimer equilibrium with a Kd of ~1 μM during SE analysis. This dimerization affinity is slightly greater than that of phosphorylated PhoB, which dimerizes with a Kd of ~5 μM. Analysis of DNA binding in vitro revealed ~2 fold lower binding affinity for PhoBZ than for phosphorylated PhoB (data not shown), supporting the notion that the longer linkers engineered into PhoBZ to avoid constraints on orientation might not be efficient in promoting optimal alignment of the C-terminal domains for DNA binding. The lower DNA-binding affinity of PhoBZ is potentially sufficient to explain the lower transcription activation observed. This suggests that the REC domain plays little if any role in interaction with polymerase, consistent with previous studies that have identified the DNA-binding domain as the locus for such interactions.62,63
The α1/α5 dimer is associated with the unphosphorylated, inactive state of PhoB. Specifically, disruption of the α1/α5 interface eliminates dimerization of unphosphorylated PhoB without decreasing either the dimerization affinity of phosphorylated PhoB or transcription activation. However, the role of the α1/α5 dimer remains unclear. Under phosphate replete conditions, when intracellular concentrations of PhoB are ~1 μM (Fig. S2), the Kd of ~400 μM would predict a negligible concentration of α1/α5 dimer. Even at 10 μM PhoB, as exists under conditions following autoinduction of PhoB, the monomeric form would predominate. However, estimates derived from dissociation constants determined in vitro are unlikely to accurately predict population distributions in vivo due to intracellular factors such as protein localization and macromolecular crowding.
It has been suggested that the α1/α5 dimer might play an inhibitory role as the C-terminal ends of the REC domains are positioned in an orientation that does not appear to allow dimerization of the DNA-binding domains for interactions with tandem DNA half-sites.23 Interestingly, and consistent with this hypothesis, under non-inducing conditions, PhoBF20D is slightly more active than PhoB. However, this level of transcription activation is minimal compared to that obtained under inducing conditions. If the α1/α5 dimer functions as an inhibitory mechanism, its contribution is small compared to the activating effect of phosphorylation-induced dimerization through the α4-β5-α5 face. Alternatively, it has been postulated that the α1/α5 dimer might facilitate interactions with histidine kinase PhoR.23 Although kinetics of activation were not examined, comparable transcription activation was observed for PhoBF20D and PhoB under inducing conditions, suggesting that disruption of the α1/α5 dimer does not substantially compromise PhoR-mediated phosphorylation.
Disruption of the α1/α5 dimer in PhoBF20D had no apparent significant physiological effects under any condition assessed. Nonetheless, the propensity to form an alternative dimer in the unphosphorylated state has been observed in both E. coli PhoB and B. subtilis PhoP,23,31,50 RRs that regulate phosphate assimilation genes. Intriguingly, the dimers are distinct, suggesting that a specific domain arrangement might not be critical to the functional role. The conserved existence of an alternative dimer across distant species suggests functional importance of this feature, although its significance has not yet been revealed in a laboratory environment. It is possible that this dimer functions in a specialized role relevant to regulation of a subset of genes that have not yet been examined, for example in promoting DNA looping that might lead to repression of transcription. Alternatively, this dimer might be involved in fine-tuning the kinetics of activation and adaptation, imparting subtle effects that fall below the sensitivity of our current analyses.
Materials and Methods
Cloning and molecular biology
For transcriptional analysis, the various PhoB constructs were cloned into the low copy number plasmid pMLBl 120.215 with protein expression driven by the lac promoter. For purification, the T7 expression systems encoded in either pJES30764 or pET-21b (Novagen, Madison, WI) were used. Native proteins were used for transcription analysis, however, for purification, PhoBF20D and PhoBR91E/E111R were expressed with a 6-histidine tag and a thrombin cleavage site. Plasmids used for expression of the various PhoB constructs were generated using one of four different cloning strategies (Tables S2 and S3 and Fig. S5).
Protein Purification
Proteins were expressed in E. coli BL-21(DE3) cells. Cells expressing PhoB, his-tagged PhoBF20D and PhoBZ were grown at 37 °C in Luria-Bertani media supplemented with 100 μg/ml ampicillin to mid-log phase, induced with 0.5 mM IPTG, and grown for an additional 2–3 h at 37 °C. Cells expressing PhoBR115D, PhoBD101R/R115D and his-tagged PhoBR91E/E111R were grown at 30 °C in Luria-Bertani media containing 1 M sorbitol, 10 mM betaine and 100 μg/ml amplicillin to mid-log phase, cooled to 20 °C prior to induction with 0.25 mM IPTG, and grown for an additional 17–20 h at 20 °C. All proteins were lysed by sonication; PhoB, his-tagged PhoBF20D and PhoBZ were lysed in a buffer containing 50 mM Tris-HCl (pH 8.0), 0.1 M NaCl, 1 mM EDTA, 0.5 mM PMSF and 2 mM β-ME while PhoBR115D, PhoBD101R/R115D and his-tagged PhoBR91E/E111R were lysed in 0.1 M sodium phosphate (pH 7.2), 5 mM EDTA and 0.5 mM PMSF. All native proteins were precipitated by the addition of solid ammonium sulfate while his-tagged proteins were not; for PhoB and PhoBR115D the concentration was 60% w/v, for PhoBD101R/R115D it was 65% w/v and for PhoBZ the final ammonium sulfate concentration was 50% w/v. Two chromatographic purification steps were used to attain a purity level of >90% as determined by SDS-PAGE analysis; the purified proteins were stored at −80 °C after rapid freezing in an ethanol dry ice bath.
PhoB and PhoBZ were applied to a 5-ml HiTrap Blue column (General Electric, Piscataway, NJ) equilibrated with 25 mM Tris-HCl and 2 mM β-ME and eluted using a linear gradient of 25 mM Tris-HCl, 2 M NaCl and 2 mM β-ME; the pH of the buffer used for PhoB was 8.0 and for PhoBZ it was 7.4. Pooled fractions containing PhoB or PhoBZ were applied to a Superdex 75 26/60 gel filtration column (General Electric, Piscataway, NJ) pre-equilibrated in 50 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 10 mM MgCl2 and 2 mM β-ME for PhoB or 25 mM Tris-HCl (pH 7.4) and 0.1 M NaCl for PhoBZ.
PhoBR115D was applied to a 5-ml Heparin HiTrap column (General Electric, Piscataway, NJ) pre-equilibrated with 10 mM sodium phosphate (pH 7.2) and 2 mM β-ME and eluted using a linear gradient of 10 mM sodium phosphate (pH 7.2), 2 M NaCl and 2 mM β-ME. Pooled fractions containing PhoBR115D were applied to a Superdex 75 26/60 gel filtration column pre-equilibrated in 25 mM sodium phosphate (pH 7.2), 0.1 M NaCl and 2 mM β-ME. The same purification protocol used for PhoBR115D was employed to purify PhoBD101R/R115D with one exception. For the heparin column purification step the flow-through was reapplied to the column and the purest fractions from that step were pooled and then loaded onto to the Superdex 75 26/60 column.
His-tagged PhoBF20D and his-tagged PhoBR91E/E111R were applied to a 5-ml HisTrap FF crude column (General Electric, Piscataway, NJ) that was pre-equilibrated with 20 mM sodium phosphate (pH 7.4), 0.5 M NaCl, 25 mM imidazole and 2 mM β-ME. The proteins were eluted using a linear gradient of 20 mM sodium phosphate (pH 7.4), 0.5 M NaCl, 0.5 M imidazole and 2 mM β-ME. Pooled fractions containing PhoBF20D were dialyzed overnight against 50 mM Tris-HCl (pH 8.0), 0.1 M NaCl and 2 mM β-ME while fractions containing PhoBR91E/E111R were dialyzed against 20 mM Tris-HCl (pH 7.5), 0.15 M NaCl and 2.5 mM CaCl2 overnight. The dialyzed materials were digested at room temperature overnight with 4–8 units of restriction grade thrombin (Novagen, San Diego, CA) to remove the histidine tag.
HPLC analysis of PhoB
Wild-type PhoB, PhoBD101R/R115D, PhoBR115D and PhoBF20D were phosphorylated at a concentration of 5 μM in a buffer containing 20 mM ammonium hydrogen phosphoramidate, 50 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 10 mM MgCl2 and 2 mM β-ME. Aliquots containing 12.5–25 μg of phosphorylated or unphosphorylated protein were injected onto a C-8 reverse-phase HPLC column (Grace-Vydac Cat# 208TP5415) pre-equilibrated with 31.5% actetonitrile, 0.1% trifluoroacetic acid. Proteins were eluted using a 60-ml gradient from 31.5% acetonitrile, 0.1% trifluoroacetic acid to 58.5% acetonitrile, 0.1% trifluoroacetic acid at a flow rate of 1 ml/min. The percentage of phosphorylated protein was calculated by dividing the area of the early eluting peak by the sum of the two peaks. For those profiles for which the two peaks could not be sufficiently resolved, a two-peak deconvolution algorithm (Hitachi Model D-7000 Chromatography Data Station Software V 4.0, Hitachi LTD, Parsippany, NJ) was applied to determine the relative amounts unphosphorylated and phosphorylated protein.
Real-time fluorescence analysis of PhoB
Phosphorylation induced fluorescence changes were monitored at 25 °C using a FluoroMax-3 spectrofluorometer (Horiba Jobin Yvon Inc. Edison, NJ). Proteins were analyzed in a 2.2 ml quartz cuvette (i.d. 1 cm × 1 cm) at a concentration of 2 μM in a buffer containing 50 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 10 mM MgCl2 and 2 mM β-ME. Phosphorylation was initiated by the addition of ammonium hydrogen phosphoramidate to a final concentration of 20 mM. Fluorescence was monitored at 345 nm after excitation at 280 nm while the reaction mixture was constantly stirred; the emission and excitation slits were set at 2 and 0.75 nm, respectively. Fluorescence profiles were fit to a single exponential decay that provided an observed phosphorylation rate constant (kobs).
Analytical ultracentrifugation
Analytical ultracentrifugation experiments were carried out in an Optima XL-I ultracentrifuge using an An-60 Ti rotor (Beckman Coulter, Fullerton, CA). Proteins were initially equilibrated in a buffer containing 50 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 10 mM MgCl2 and 5 mM TCEP via exhaustive dialysis. The dialyzed proteins were phosphorylated at concentrations ranging from 5–20 μM by the addition of phosphoramidate (20 mM) and subsequent incubation at room temperature for 1–3 h. Buffer density, buffer viscosity and the partial specific volumes of the proteins were calculated using the program SEDNTERP.65 The contribution of phosphoramidate to the buffer density and viscosity was not accounted for due to its low concentration. For all experiments, the protein gradients were monitored using the absorbance optical system (280 nm). SV runs were conducted in epon charcoal-filled double-sector centerpieces while SE runs were carried out in six-channel epon charcoal-filled centerpieces; quartz windows were used for both. SV experiments were conducted at 24 °C and the samples were spun at 50k RPM. Approximately 50 SV scans were used to calculate a c(s) distribution that was generated using the program SEDFIT (v94).44 During the c(s) analysis, the frictional ratio and meniscus position were treated as floating parameters. After optimization of these parameters, the final distribution was calculated using a resolution setting of 200 and a confidence interval of 0.8. For SE experiments, three different protein concentrations (5, 10 and 20 μM) at two different speeds (24 and 30k RPM) were analyzed at 15 °C. The program WINMATCH (v0.99) was used to ensure the samples had reached equilibrium. A total of 6 different SE profiles were globally fit to either the singles species model or the monomer-dimer self association model using the program SEDPHAT (v 4.0).66 Error limits, which represent 1 standard deviation, were determined using F-statistics.
Fluorescence anisotropy
An oligonucleotide was designed to contain the pho box found within the promoter region of phoB (Fig. S1) and adjacent base pairs that contact PhoB in the crystal structure of the PhoB-DNA complex.37 This oligonucleotide was synthesized by Integrated DNA Technologies (Caorville, IL) with a primary amino group attached to the 5′ end of the non-coding strand via a 6-carbon linker. A fluorescent moiety, TAMRA (carboxytetramethylrhodamine), was attached to the oligonucleotide via the primary amino group. Experiments were conducted on a FluoroMax-3 spectrofluorometer (Horiba Jobin Yvon Inc. Edison, NJ) in a buffer containing 50 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 10 mM MgCl2 and 2 mM β-ME, with 10 nM TAMRA-labeled oligonucleotide. For titrations with phosphorylated proteins, 20 mM ammonium hydrogen phosphoramidate was also present, and proteins (5–20 μM) were phosphorylated in titration buffer containing phosphoramidate for 30–90 min at room temperature. Fluorescence anisotropy was monitored using an excitation wavelength of 555 nm and an emission wavelength of 585 nm; the excitation and emission slits were set at 3 and 10 nm, respectively. Titrations were carried out at 24 °C in a 1.5-ml cuvette (i.d. 0.4 cm × 1 cm) with constant stirring. The observed anisotropy (robs) represents the average of three readings taken after a 1-min incubation period. The binding isotherm was fit to the following model:
where Kd is the dissociation constant, P is the protein concentration for each titration step, Dt is the total DNA concentration, n is the stochiometry and rf and rb are the anisotropies of the free and protein-bound forms of DNA, respectively. During the fitting of the data, Kd, rf and rb were treated as floating parameters while n and Dt were fixed. The stoichiometry was fixed at two moles of protein per mole of DNA since this oligonucleotide contains one pho box, which accommodates two protein molecules.
Transcriptional activation assays
PhoB proteins were expressed under the control of the lac promoter in a phoB deletion strain (Genobase ID: JW038945). Cells were grown overnight at 37 °C in 3 ml of Luria-Bertani media supplemented with 100 μg/ml ampicillin and 1% glucose. Stationary phase cells were harvested by centrifugation (2450 g), washed twice with MOPS minimal media67 (without phosphate) and resuspended in 3 ml of the same solution. These cells were used to inoculate 3-ml cultures of MOPS minimal media at a final optical density at 595 nm of ~0.04 AU. The cultures were supplemented with 100 μg/ml ampicillin, either 5 mM (high phosphate conditions) or 0.2 mM (low phosphate conditions) dibasic potassium phosphate and IPTG at concentrations ranging from 2 to 100 μM. Cells were grown at 37 °C with vigorous shaking to an optical density of ~0.7 AU. Cells were harvested from 0.5-ml aliquots by centrifugation (18000 g) and cell pellets were stored at −20 °C. Frozen cell pellets were suspended in 0.5 ml of buffer containing 0.5 M Tris-HCl (pH 8.2) and 1 mM MgCl2 and lysed by the addition of 18 μl of 0.1% SDS and 36 μl of chloroform. After vigorous shaking the cell lysate was clarified by centrifugation (18000 g). In a 96-well plate, 100 μl of clarified supernatant and 25 μl of a solution containing 4 mM p-nitrophenyl phosphate, 0.5 M Tris-HCl (pH 8.2) and 1 mM MgCl2 were mixed just prior to analysis on a Thermo Scientific Varioskan plate reader (Thermo Fisher Scientific, Inc., Waltham, MA). The absorbance change at 420 nm was monitored over a 45 min period. Relative alkaline phosphatase activity is expressed as the linear rate of change in absorbance; only data points below 1 AU were included. The final activity was corrected for differences in cell density. For cells expressing two PhoB mutants, the same method was followed except that additional PhoB mutant proteins were under the control of the pBAD promoter and induced by 0.02% arabinose to match the expression level of the first mutant protein.
Supplementary Material
Acknowledgments
We thank J. Gao and T. Wu for assistance in plasmid construction and protein purification and J. Guhaniyogi and Y. Tao for synthesis of phosphoramidate. This work was supported by a grant from the National Institutes of Health (R37GM047958). AMS is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- AP
alkaline phosphatase
- AU
absorbance units
- βME
β-mercaptoethanol
- EDTA
ethylenediaminetetraacetic acid
- HK
histidine kinase
- HPLC
high performance liquid chromatography
- MOPS
3(N-morpholino)propanesulfonic acid
- IPTG
isopropyl-l-thio-β-D-galactopyranoside
- PMSF
phenylmethylsulfonyl fluoride
- REC
receiver
- RR
response regulator
- TCEP
Tris(2-Carboxyethyl) phosphine
- TCS
two-component system
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SE
sedimentation equilibrium
- SV
sedimentation velocity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hoch JA, Silhavy TJ, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology Press; 1995. [Google Scholar]
- 2.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 4.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 5.Peterson CN, Mandel MJ, Silhavy TJ. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J Bacteriol. 2005;187:7549–7553. doi: 10.1128/JB.187.22.7549-7553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 7.Beier D, Gross R. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol. 2006;9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Barrett JF, Hoch JA. Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob Agents Chemother. 1998;42:1529–1536. doi: 10.1128/aac.42.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett JF, Goldschmidt RM, Lawrence LE, Foleno B, Chen R, Demers JP, Johnson S, Kanojia R, Fernandez J, Bernstein J, Licata L, Donetz A, Huang S, Hlasta DJ, Macielag MJ, Ohemeng K, Frechette R, Frosco MB, Klaubert DH, Whiteley JM, Wang L, Hoch JA. Antibacterial agents that inhibit two-component signal transduction systems. Proc Natl Acad Sci USA. 1998;95:5317–5322. doi: 10.1073/pnas.95.9.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macielag MJ, Goldschmidt R. Inhibitors of bacterial two-component signalling systems. Expert Opin Investig Drugs. 2000;9:2351–2369. doi: 10.1517/13543784.9.10.2351. [DOI] [PubMed] [Google Scholar]
- 11.Stephenson K, Hoch JA. Developing inhibitors to selectively target two-component and phosphorelay signal transduction systems of pathogenic microorganisms. Curr Med Chem. 2004;11:765–773. doi: 10.2174/0929867043455765. [DOI] [PubMed] [Google Scholar]
- 12.Rasko DA, Moreira CG, Li de R, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 14.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulrich LE, Zhulin IB. MiST: a microbial signal transduction database. Nucl Acids Res. 2007;35:D386–D390. doi: 10.1093/nar/gkl932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo H, Nakagawa A, Nishihira J, Nishimura Y, Mizuno T, Tanaka I. Escherichia coli positive regulator OmpR has a large loop structure at the putative RNA polymerase interaction site. Nat Struct Biol. 1997;4:28–31. doi: 10.1038/nsb0197-28. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Hackert E, Stock AM. The DNA-binding domain of OmpR: crystal structure of a winged-helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Hackert E, Stock AM. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997;269:301–312. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 19.Park S, Meyer M, Jones AD, Yennawar HP, Yennawar NH, Nixon BT. Two-component signaling in the AAA+ ATPase DctD: binding Mg2+ and BeF3− selects between alternate dimeric states of the receiver domain. FASEB J. 2002;16:1964–1966. doi: 10.1096/fj.02-0395fje. [DOI] [PubMed] [Google Scholar]
- 20.Lee SY, De La Torre A, Yan D, Kustu S, Nixon BT, Wemmer DE. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 2003;17:2552–2563. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Carlo S, Chen B, Hoover TR, Kondrashkina E, Nogales E, Nixon BT. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–95. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci. 2007;32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachhawat P, Swapna GV, Montelione GT, Stock AM. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure. 2005;13:1353–1363. doi: 10.1016/j.str.2005.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiedler U, Weiss V. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of the receiver modules. EMBO J. 1995;14:3696–3705. doi: 10.1002/j.1460-2075.1995.tb00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCleary WR. The activation of PhoB by acetylphosphate. Mol Microbiol. 1996;20:1155–1163. doi: 10.1111/j.1365-2958.1996.tb02636.x. [DOI] [PubMed] [Google Scholar]
- 26.Jeon Y, Lee YS, Han JS, Kim JB, Hwang DS. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J Biol Chem. 2001;276:40873–40879. doi: 10.1074/jbc.M104855200. [DOI] [PubMed] [Google Scholar]
- 27.Robinson VL, Wu T, Stock AM. Structural analysis of the domain interface in DrrB, a response regulator of the OmpR/PhoB subfamily. J Bacteriol. 2003;185:4186–4194. doi: 10.1128/JB.185.14.4186-4194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toro-Roman A, Mack TR, Stock AM. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the α4-β5-α5 face. J Mol Biol. 2005;349:11–26. doi: 10.1016/j.jmb.2005.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toro-Roman A, Wu T, Stock AM. A common dimerization interface in bacterial response regulators KdpE and TorR. Protein Sci. 2005;14:3077–388. doi: 10.1110/ps.051722805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. Structure of BeF3−-modified response reuglator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure. 2007;15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 31.Solà M, Gomis-Rüth FX, Serrano L, González A, Coll M. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J Mol Biol. 1999;285:675–687. doi: 10.1006/jmbi.1998.2326. [DOI] [PubMed] [Google Scholar]
- 32.Wanner BL. Gene regulation by phosphate in enteric bacteria. J Cell Biochem. 1993;51:47–54. doi: 10.1002/jcb.240510110. [DOI] [PubMed] [Google Scholar]
- 33.Wanner BL. Signal transduction and cross regulation in the Escherichia coli phosphate regulon by PhoR, CreC, and acetyl phosphate. In: Hoch JA, Silhavy TJ, editors. Two-Component Signal Transduction. American Society for Microbiology Press; Washington, D.C: 1995. pp. 203–221. [Google Scholar]
- 34.Wanner BL. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low J, Magasanik KBB, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella. American Society for Microbiology Press; Washington, D.C: 1996. pp. 1357–1381. [Google Scholar]
- 35.Lamarche MG, Wanner BL, Crepin S, Harel J. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev. 2008;32:461–473. doi: 10.1111/j.1574-6976.2008.00101.x. [DOI] [PubMed] [Google Scholar]
- 36.Okamura H, Hanaoka S, Nagadoi A, Makino K, Nishimura Y. Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box. J Mol Biol. 2000;295:1225–1236. doi: 10.1006/jmbi.1999.3379. [DOI] [PubMed] [Google Scholar]
- 37.Blanco AG, Sola M, Gomis-Ruth FX, Coll M. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure. 2002;10:701–713. doi: 10.1016/s0969-2126(02)00761-x. [DOI] [PubMed] [Google Scholar]
- 38.Solà M, Drew DL, Blanco AG, Gomis-Rüth FX, Coll M. The cofactor-induced pre-active conformation in PhoB. Acta Crystallogr D Biol Crystallogr. 2006;62:1046–1057. doi: 10.1107/S0907444906024541. [DOI] [PubMed] [Google Scholar]
- 39.Arribas-Bosacoma R, Kim SK, Ferrer-Orta C, Blanco AG, Pereira PJ, Gomis-Rüth FX, Wanner BL, Coll M, Solà M. The X-ray crystal structures of two constitutively active mutants of the Escherichia coli PhoB receiver domain give insights into activation. J Mol Biol. 2007;366:626–641. doi: 10.1016/j.jmb.2006.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson VL, Buckler DR, Stock AM. A tale of two components: a novel kinase and a regulatory switch. Nat Struct Biol. 2000;7:628–633. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- 41.Bent CJ, Isaacs NW, Mitchell TJ, Riboldi-Tunnicliffe A. Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states. J Bacteriol. 2004;186:2872–2879. doi: 10.1128/JB.186.9.2872-2879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bachhawat P, Stock AM. Crystal structures of the receiver domain of the response regulator PhoP from Escherichia coli in the absence and presence of the phosphoryl analog beryllofluoride. J Bacteriol. 2007;189:5987–5995. doi: 10.1128/JB.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedland N, Mack TR, Yu M, Hung LW, Terwilliger TC, Waldo GS, Stock AM. Domain orientation in the inactive response regulator Mycobacterium tuberculosis MtrA provides a barrier to activation. Biochemistry. 2007;46:6733–6743. doi: 10.1021/bi602546q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez LJ, Woolfson DN, Alber T. Buried polar residues and structural specificity in the GCN4 leucine zipper. Nat Struct Biol. 1996;3:1011–1018. doi: 10.1038/nsb1296-1011. [DOI] [PubMed] [Google Scholar]
- 48.Ellison DW, McCleary WR. The unphosphorylated receiver domain of PhoB silences the activity of its output domain. J Bacteriol. 2000;182:6592–6597. doi: 10.1128/jb.182.23.6592-6597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harlocker SL, Bergstrom L, Inouye M. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J Biol Chem. 1995;270:26849–26856. doi: 10.1074/jbc.270.45.26849. [DOI] [PubMed] [Google Scholar]
- 50.Birck C, Chen Y, Hulett FM, Samama JP. The crystal structure of the phosphorylation domain in PhoP reveals a functional tandem association mediated by an asymmetric interface. J Bacteriol. 2003;185:254–261. doi: 10.1128/JB.185.1.254-261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Birck C, Samama JP, Hulett FM. Residue R113 is essential for PhoP dimerization and function: a residue buried in the asymmetric PhoP dimer interface determined in the PhoPN three-dimensional crystal structure. J Bacteriol. 2003;185:262–273. doi: 10.1128/JB.185.1.262-273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simonovic M, Volz K. A distinct meta-active conformation in the 1.1-Å resolution structure of wild-type apoCheY. J Biol Chem. 2001;276:28637–28640. doi: 10.1074/jbc.C100295200. [DOI] [PubMed] [Google Scholar]
- 53.Kern D, Volkman BF, Luginbuhl P, Nohaile MJ, Kustu S, Wemmer DE. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;40:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- 54.Dyer CM, Quillin ML, Campos A, Lu J, McEvoy MM, Hausrath AC, Westbrook EM, Matsumura P, Matthews BW, Dahlquist FW. Structure of the constitutively active double mutant CheYD13K Y106W alone and in complex with a FliM peptide. J Mol Biol. 2004;342:1325–1335. doi: 10.1016/j.jmb.2004.07.084. [DOI] [PubMed] [Google Scholar]
- 55.Guhaniyogi J, Robinson VL, Stock AM. Crystal structures of beryllium fluoride-free and beryllium fluoride-bound CheY in complex with the conserved C-terminal peptide of CheZ reveal dual binding modes specific to CheY conformation. J Mol Biol. 2006;359:624–645. doi: 10.1016/j.jmb.2006.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dyer CM, Dahlquist FW. Switched or not? The structure of unphosphorylated CheY bound to the N terminus of FliM. J Bacteriol. 2006;188:7354–7363. doi: 10.1128/JB.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuster M, Silversmith RE, Bourret RB. Conformational coupling in the chemotaxis response regulator CheY. Proc Natl Acad Sci USA. 2001;98:6003–6008. doi: 10.1073/pnas.101571298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 59.Jiang M, Bourret RB, Simon MI, Volz K. Uncoupled phosphorylation and activation in bacterial chemotaxis: The 2.3 Å structure of an aspartate to lysine mutant at position 13 of CheY. J Biol Chem. 1997;272:11850–11855. doi: 10.1074/jbc.272.18.11850. [DOI] [PubMed] [Google Scholar]
- 60.Walthers D, Tran VK, Kenney LJ. Interdomain linkers of homologous response regulators determine their mechanism of action. J Bacteriol. 2003;185:317–324. doi: 10.1128/JB.185.1.317-324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mattison K, Oropeza R, Kenney LJ. The linker region plays an important role in the inter-domain communication of the response regulator OmpR. J Biol Chem. 2002;277:32714–32721. doi: 10.1074/jbc.M204122200. [DOI] [PubMed] [Google Scholar]
- 62.Makino K, Amemura M, Kim SK, Nakata A, Shinagawa H. Role of the σ70 subunit of RNA polymerase in transcription activation by activator protein PhoB in Escherichia coli. Genes Dev. 1993;7:149–160. doi: 10.1101/gad.7.1.149. [DOI] [PubMed] [Google Scholar]
- 63.Slauch JM, Russo FD, Silhavy TJ. Suppressor mutations in rpoA suggest that OmpR controls transcription by direct interaction with the alpha subunit of RNA polymerase. J Bacteriol. 1991;173:7501–7510. doi: 10.1128/jb.173.23.7501-7510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCleary WR, McBride MJ, Zusman DR. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990;172:4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton J, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 66.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 67.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.