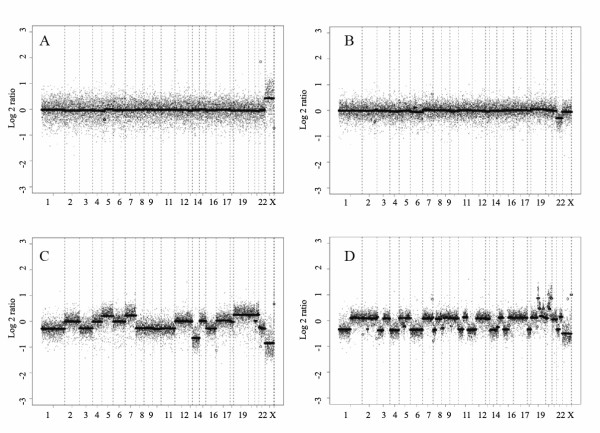
Figure 1.
Representative array CGH profiles of meningiomas. Segmentation analysis of array CGH results generated from Agilent cDNA arrays using four representative sporadic solitary meningiomas is shown: A: Benign meningioma (68B) without imbalanced chromosome segments (ICS). The X chromosome imbalance is due to use of a sex-mismatched standard DNA as a positive control; B: Meningioma with chromosome 22 deletion as the only imbalance event (42A); C: Atypical meningioma with both gain and loss ICS (43A); D: Malignant meningioma with many ICS and focal amplification (129M). X-axis: clones are ordered from chromosome 1 to 22, X and Y and within each chromosome, clones are arranged following their physical map order from short arm telomere to long arm telomere. Y-axis: log2 ratio (test/control) of array CGH signal for each individual probe (scattered dots) and for segments of consistent dosage (solid lines) defined by binary segmentation. The baseline (no copy number change) is 0; segments above the baseline indicate gain of copy number and segments below the baseline indicate loss of copy number.