Abstract
Introduction
Interleukin (IL)-18 plays an important dual role in Th1 polarization and viral clearance, as well as in the development of liver fibrosis. Single-nucleotide promoter polymorphisms influence the transcription of IL-18 mRNA. Promoter polymorphisms are linked to delayed virus clearance and disease susceptibility in many diseases. However, there is no information about their role in hepatitis C virus (HCV) infection.
Aim
To investigate the association between −607 or −137 polymorphism with susceptibility and severity of HCV infection.
Patients and methods
Two hundred and four serologically proven patients with chronic HCV infection and 350 matched healthy controls were included in this study. Patients were segregated in 2 groups: group A with mild liver disease and group B with severe liver disease on the basis of histological activity index (HAI ≤5 or >5) and hepatic fibrosis score (≤2 or >2). IL-18 promoter genotyping was performed with sequence-specific primers.
Results
There was no significant difference in the frequencies of −607 and −137 allelic distribution in patients and controls. The −607 A/A allele was more common in group A patients with mild liver disease than in patients with severe liver disease on the basis of HAI (38.6% vs. 21%, P = 0.05; odds ratio [OR] = 0.424, confidence interval [CI] = 0.233–0.773; R2 = 0.631) and stage of fibrosis (38.7% vs. 16.7%, P = 0.008; OR = 0282, CI = 0.134–0.596; R2 = 0.434).
Conclusions
IL-18 promoter polymorphism at −607 position with A/A allele is a potential protective marker, as it is associated with milder liver disease in patients with chronic HCV infection.
Keywords: IL-18, Hepatitis C, Polymorphism, HAI, Fibrosis
Introduction
About one-third of patients infected with chronic hepatitis C virus (HCV) develop cirrhosis and hepatocellular carcinoma (HCC) [1]. The rate at which the liver disease develops varies from one individual to other. There is sufficient evidence to suggest that the disease progression is multifactorial, including viral, host immunological, and genetic factors. The predominant genes involved in HCV disease progression and persistence are those that are involved in the replication and mediation of immunological response. Cytokines play important role in differentiation, maturation, and functional activation of immune cells.
Interindividual variations in cytokine production, and polymorphisms in cytokine and/or their receptor genes, may directly influence the outcome of cytokine-based immunotherapy. Genetic variation exerts a major influence on susceptibility and progression of infectious diseases [2]. The association of tumor necrosis factor (TNF)-β and vitamin D receptor gene polymorphisms was also linked to viral persistence and severity of liver disease in HCV patients [3].
Interleukin (IL)-18, also called interferon (IFN)-γ–inducing factor, is an obligatory cytokine for IFN-γ production [4] and plays a key role in the induction of Th1 responses [5]. Pro-IL-18 is a biologically inactive precursor that is produced during the acute immune response by monocytes, macrophages, and immature dendritic cells. It is activated intracellularly by caspase-1 to induce IFN-γ and TNF-α secretion. It also enhances cytotoxicity of natural killer cells and FasL expression.
IFN-α exerts its anti-inflammatory action by the induction of IL-18-binding protein (IL-18BP). Increased IL-18 production is neutralized by IL-18BP in chronic HCV infection, and this neutralization is crucial for the regulation of inflammation and fibrosis development [4]. IFN therapy increases plasma IL-18BP levels by 3- to 24-fold within 24 h following the institution of therapy [6].
Asakawa et al. [7] showed that elevated level of IL-18 receptors was a significant predictor of poor outcome of IFN therapy in HCC patients. In addition to this, animal models have also shown lethal lung injury due to IL-18 induction [8], thereby suggesting that IL-18 is an important factor for the development of inflammation and liver disease.
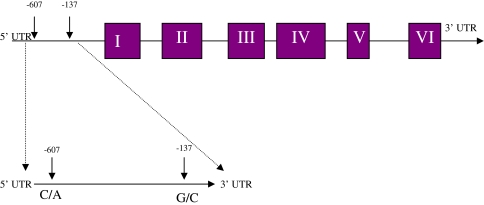
Earlier studies suggest that IL-18 promoter polymorphism at −607 and −137 positions (Fig. 1) is associated with susceptibility to HBV infection [9, 10]. The IL-18BP promoter resides within 1.6-kb DNA upstream of the first exon and includes at least six regulatory elements. The basal promoter is a γ-activated sequence (GAS), which is proximal to the transcription start site (base 1), followed by an IFN regulatory factor 1 response element (IRF-E) and 2 CCAAT/enhancer binding protein β (C/EBPβ) sites, all of which are essential for the basal promoter activity. Furthermore, GAS and IRF-E are essential for IFN-γ-induced transcription. Decreased HBV replication was also associated with −607 AA allele polymorphism. IL-18 gene polymorphism at −607 and −137 positions has been shown to be associated with Crohn’s disease [11], cardiovascular diseases [12], and infectious diseases such as human immunodeficiency virus (HIV) infection [13]. However, there is no information on the relevance and significance of IL-18 promoter polymorphism at −607 and −137 positions in HCV. This study is designed to investigate the frequency of polymorphism at −607 and −137 positions and its association with liver disease severity in HCV-infected patients.
Fig. 1.
Schematic representation of IL-18 gene with promoter region and regions analyzed for promoter polymorphisms
Patients and methods
Patients
Two hundred and four patients with histological proven chronic HCV infection were studied. The inclusion criteria were evidence of chronic hepatitis in liver biopsy, proof of the presence of HCV-RNA by RT-PCR, and HCV-RNA quantification by bDNA assay (Chiron Corporation, Emeryville, CA). Patients were excluded if they had hepatitis B virus (HBV) or HIV infection; history of heavy alcohol consumption (>80 g/day for >5 years); were positive for antinuclear or antismooth muscle antibody (in 1:80 dilution); and had autoimmune liver disease, thyroid disease, diabetes mellitus, or malaria.
Controls
A total of 350 unrelated healthy adult subjects, with no previous history of liver diseases and negative for HBV and HCV infections, were included as controls. Patients and controls were prospectively matched for ethnic group.
The institutional ethical committee approved the study protocol. An informed consent was obtained for enrolling the patients. Clinical and biochemical assessments of the patients were performed according to the study protocol.
Histological examination
The histological activity index (HAI), a marker of the inflammatory activity in the liver, and the stage of hepatic fibrosis, an indication of the fibrosis in the liver, were assessed in individual biopsy specimens in a blinded manner by using the modified Knodell scoring system [14]. Based on the HAI and hepatic fibrosis scores, the patients were divided into 2 groups: group A, patients with mild liver disease (HAI ≤ 5 and fibrosis score ≤2) and group B, patients with severe liver disease (HAI > 5 and fibrosis score >2).
Serology
Blood from the patients was collected and the serum and peripheral blood mononuclear cells were separated and stored in aliquots at −80°C. Antibodies for HCV were detected by the third-generation ELISA kit. Routine hematological and biochemical tests were performed in each patient.
HCV-RNA detection
After confirmation of the presence of antibodies against HCV, HCV-RNA was extracted from 140 μl of serum sample by using viral RNA isolation kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions. The highly conserved 50-noncoding region of HCV genome was amplified as described earlier [15], with slight modifications.
HCV genotyping
The genotype of the amplified cDNA was determined by the principle of reverse hybridization using the line probe assay (INNO-LiPA HCV II Kit, InnogeneticsQ2, Ghent, Belgium).
DNA extraction and PCR-based genotyping of allelic variants
Genomic DNA was extracted from the whole blood by using sodium per chlorate method [16]. Genotyping of IL-18 promoter region at −607 and −137 positions was performed with PCR-SSP [17]. One sequence-specific primer (C/A) at −607 position and the common reverse primer were included in every reaction mixture at a concentration of 10 nmol. In addition, 1.25 μl of the internal positive control primer was added to the reaction mixture. Internal positive amplification control was used to amplify a 301-bp fragment (for position −607) or 446-bp fragment (for position −137) covering the polymorphic site (Table 1).
Table 1.
Sequence-specific primers for C/A and G/C alleles and their PCR product sizes for positions −607 and −137
| Primer sequence | Product size (bp) | |
|---|---|---|
| Sequence-specific primers for C/A allele at position −607 | ||
| Common reverse primer | 5′-TAACCTCATTCAGGACTTCC-3′ | |
| Sequence-specific forward primers 1 | 5′-GTTGCAGAAAGTGTAAAAATTATTAC-3′ | 196 |
| Sequence-specific forward primers 2 | 5′-GTTGCAGAAAGTGTAAAAATTATTAA-3′ | 196 |
| Control forward primer | 5′-CTTTGCTATCATTCCAGGAA-3′ | 301 |
| Sequence-specific primers for G/C allele at position −137 | ||
| Common reverse primer | 5′-AGGAGGGCAAAATG CACTGG-3′ | |
| Sequence-specific forward primers 1 | 5′-CCCCAACTTTTACGGAAGAAAAG-3′ | 261 |
| Sequence-specific forward primers 2 | 5′-CCCCAACTTTTACGGAAGAAAAC-3′ | 261 |
| Control forward primer | 5′-CCAATAGGACTGATTAT TCCGCA-3′ | 446 |
The reaction was performed in a final volume of 25 μl consisting of 2.5 μl of 10× PCR buffer, 0.5 μl of 10 mmol of dNTP, 1.5 μl of 25 mmol MgCl2, 20 ng of genomic DNA, and 0.5 units of Taq polymerase. Reactions were carried out in an ABI thermocycler with denaturation for 2 min at 94°C; followed by 7 cycles of 94 for 20 s, 64 for 40 s, and 72 for 40 s; and 25 cycles of 94 for 20 s, 57 for 40 s, 72 for 40 s, and 72 for 7 min. PCR products were visualized by 2% agarose gel electrophoresis stained by ethidium bromide.
Statistical analysis
Allelic frequencies were compared between patients with chronic HCV infection and controls by χ2 test. Categorical variables were analyzed with χ2 or Fisher’s exact test. Two-sample t tests were used to compare means for continuous variables, and nonparametric test and Wilcoxon Mann–Whitney U test were used for the comparison of median values for nonnormally distributed continuous variables. Univariate analysis was used to assess associations between the various allelic variants and severity of liver disease.
The Hardy–Weinberg equilibrium was tested by comparing expected and observed genotype frequencies by χ2 test. The distribution of genotypes between the patients and the healthy controls was compared by contingency table analysis.
For all tests, a 2-tailed P < 0.05 was considered significant. The analysis was performed with statistical software SPSS 12.0 (SPSS Inc., Chicago, IL).
Results
Demographic profile
Two hundred four patients with chronic HCV infection were studied. The baseline characteristics of the patients and healthy controls in the study group are summarized in Table 2. The most common source of HCV infection was blood transfusion.
Table 2.
Demographic profile of patients with chronic HCV infection and healthy controls
| Patients (n = 204) | Healthy controls (n = 350) | |
|---|---|---|
| Age, mean ± SD (years) | 45 ± 12 | 32 ± 10 |
| Male: Female | 126:78 | 240:110 |
| ALT (IU/l) | 102 ± 56 | 26.95 ± 4.6 |
| Serum albumin (g/dl) | 3.8 ± 0.4 | 3.5 ± 0.5 |
| HCV genotype (n = 204) | ||
| 1 | 33 (16.17%) | – |
| 2 | 4 (1.47%) | – |
| 3 | 156 (76.4%) | – |
| 4 | 11 (5.39%) | – |
| Liver biopsy | ||
| Fibrosis | ||
| Mild | 83 (40.6%) | – |
| Severe | 120 (59%) | – |
| HAI | ||
| Mild | 93 (45.5%) | – |
| Severe | 111 (54.4%) | – |
| Mode of acquisition of HCV | ||
| Blood transfusion | 73 (35%) | – |
| Needle-stick injury | 38 (19%) | – |
| Medical procedure | 58 (28%) | – |
| Unclear | 35 (18%) | – |
Viral genotype
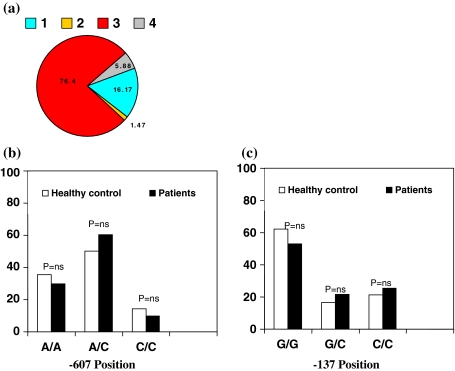
HCV genotypes could be determined in 204 (100%) patients. The predominant viral genotype was 3a/3b. One hundred forty-eight (76.6%) patients had genotype 3, 31 (16.06%) had genotype 1, 3 (1.47%) had genotype 2, and 11 (5.39%) had genotype 4. No significant difference was observed in immune gene polymorphism and patients with different viral genotypes (Fig. 2a).
Fig. 2.
a HCV genotype distribution. b and c IL-18 polymorphisms and allocation of alleles at −607 and −137 positions in healthy controls and HCV patients
Correlation of IL-18 promoter polymorphism with susceptibility to HCV infection
We studied IL-18 promoter polymorphism at −607C/A and −137G/C positions with susceptibility to HCV infection. The allele frequencies were compared between the patients and control groups. There was no significant difference in the distribution of all the alleles at −607 and −137 positions in patients and controls (Fig. 2b).
Correlation of IL-18 promoter polymorphism at −607 position with severity of HCV-related liver disease
To study whether IL-18 gene polymorphisms influence the course of HCV-related liver disease, allelic distribution was compared between group A with mild liver disease and group B patients with severe liver disease.
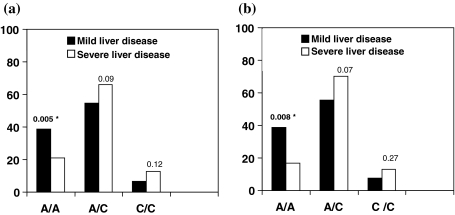
The frequency of A/A allele at −607 position was more in group A patients with mild liver disease than in group B patients with severe liver disease on the basis of HAI (38.6% vs. 21%, P = 0.005; odds ratio [OR] = 0.424, confidence interval [CI] = 0.233–0.773; R2 = 0.631) and fibrosis (38.7% vs. 16.7%, P = 0.008; OR = 0282, CI = 0.134–0.596; R2 = 0.434), whereas the frequency of A/C allele at −607 position was not associated with HAI (54.7% vs. 66.0%, P = 0.09) and fibrosis (55.6% vs. 70.3%, P = 0.07) and C/C allele frequency was also significantly not recurrent with HAI (6.6% vs. 12.8%, P = 0.12) and fibrosis (7.69% vs. 12.96%, P = 0.271) (Fig. 3).
Fig. 3.
Frequency of IL-18 polymorphism alleles at −607 position in patients with mild and severe liver diseases. a Distribution of patients with mild and severe liver diseases on the basis of HAI stage. b Distribution of patients with mild and severe liver diseases on the basis of fibrosis score
Correlation of IL-18 promoter polymorphism at −137 position with severity of HCV-related liver disease
There was no significant difference in allelic distribution at −137 positions between patients with mild and severe liver diseases. Distribution of G/G, G/C, and C/C alleles were same in patients with mild and severe liver diseases; G/G allele and HAI (55.6% vs. 51.2%, P = 0.531) and fibrosis (51% vs. 50.7%, P = 0.970), G/C allele and HAI (24.4% vs. 19.5%, P = 0.38) and fibrosis (22.4% vs. 21.7%, P = 0.913), and C/C allele and HAI (20% vs. 29.2%, P = 0.12) and fibrosis (28.5% vs. 27.5%, P = 0.885).
Our results indicate that IL-18 gene polymorphism is associated with severity of disease but not susceptibility of HCV disease.
Discussion
IL-18 induces the expression of chemokines, IL-8, IL-2, IFN-γ, and T cells. Molecular mechanism for the production of IL-18 is mediated by TRAF-6 (TNF receptor associated factor 6). Interleukin receptor activates myeloid differentiation factor-88 (MyD88) and IL-1 receptor-associated kinase (IRAK), with the activation of NF-κB. This activation leads to the synthesis of proinflammatory genes, such as iNOS (inducible nitric oxide) and IFN-γ [18, 19]. IL-18 also enhances proinflammatory activity by inducing matrix metalloproteinases, which are crucial for pathological chemotaxis of immune cells to target tissues [20]. It also enhances hepatic injury and the level of IL-2, which is a marker of inflammation [21] (Fig. 1).
To study whether susceptibility or severity of HCV infection is linked to IL-18, a promoter of immune gene polymorphism, patients with chronic HCV infection were compared with healthy controls. Single-nucleotide polymorphisms of IL-18, transversion C to A at position −607, and a transversion G to C at position−137 modify two transcription binding sites that influence the quantity of transcribed IL-18 mRNA [17]. In the present study, the frequencies of all the alleles at −607 and −137 positions were alike between patients and controls. However, when patients were divided into two groups on the basis of HAI and fibrosis, A/A allele at −607 position was more frequent in patients with mild liver disease than in patients with severe liver disease on the basis of HAI (P = 0.005) and stage of fibrosis (P = 0.008). This association was not observed with A/C or C/C allele at −607 position. On the other hand, there was no significant difference in the allelic distribution at −137 positions between patients with mild and severe liver diseases; there was no association between G/G allele and HAI (P = 0.531) and fibrosis (P = 0.970), G/C allele and HAI (P = 0.38) and fibrosis (P = 0.913), and C/C allele and HAI (P = 0.12) and fibrosis (P = 0.885).
Serum IL-18 and HCV-RNA titers were highly correlated with IL-18 levels in patients treated with IFN and ribavirin combination therapy [22]. Recombinant IL-18 can also be given safely in biologically active doses to patients with advanced cancer [23]. It has been shown that IL-18 levels correlate with the serum alanine aminotransferase and aspartate aminotransferase activities, markers of hepatic injury [21]. Of the 204 patients, 54.4% had severe and 45.5% had mild liver diseases on the basis of HAI (>5 or ≤5), and 59% had severe and 40.6% had mild liver diseases (>2 or ≤2) on the basis of the stage of hepatic fibrosis. In the present study, HCV genotype 3-infected patients had higher HAI (55.4% vs. 38.7%) and fibrosis (53.1% vs. 32.2%) than genotype 1-infected patients, although the differences were not significant. This nonsignificance could be due to fewer patients in genotype 1 group than in genotype 3 group.
In different populations, IL-18 gene polymorphism at either −607 or −137 position, or both, has been shown to influence the severity of many diseases, for example, Crohn’s disease [11], cardiovascular diseases [12], and infectious diseases such as HIV infection [13] (Table 3). Polymorphisms of IL-18 gene promoter have also been shown to be closely associated with the susceptibility to chronic HBV infection [9]. Our results suggest that there is no correlation between the susceptibility to HCV infection and IL-18 gene promoter polymorphism. However, there was a significant correlation with disease severity.
Table 3.
Association of IL-18 promoter polymorphism in populations with different diseases and disease severity
| Disease | Position | Population | Outcome | Reference |
|---|---|---|---|---|
| HIV | −607 | Brazilian | Associated | [13] |
| Rheumatoid arthritis | −607, −137 | 309 Polish | Associated | [24] |
| Type 1 diabetes | −607 | 112 Iranians | No association | [25, 26] |
| Allergic rhinitis | −607 | 160 Korea | Associated | [27] |
| Crohn’s disease | −607, −137 | 210 German | Association | [11] |
| Behcet’s disease | −607, −137 | 98 Korea | No association | [28] |
| Sarcoidosis | −607, −137 | 161 Japanese | No association | [29] |
| Periodontitis | −607 | 123 German | No association | [30] |
| Cardiovascular disease | −137 | 1288 French | Associated | [12] |
| Atopic eczema | −137 | 225 German | Associated | [31] |
| Inflammatory bowel disease | −137 | 178 Japan | Associated | [32] |
In summary, IL-18 gene polymorphism does not have a strong correlation with susceptibility to chronic HCV infection. On the other hand, study of this polymorphism is helpful since we observed that A/A allele at −607 position of IL-18 gene promoter is associated with milder liver disease.
References
- 1.Lechmann M, Liang TJ. Vaccine development for hepatitis C. Semin Liver Dis. 2000;20(2):211–226. doi: 10.1055/s-2000-9947. [DOI] [PubMed] [Google Scholar]
- 2.Hill AV. Genetics and genomics of infectious disease susceptibility. Br Med Bull. 1999;55(2):401–413. doi: 10.1258/0007142991902457. [DOI] [PubMed] [Google Scholar]
- 3.Goyal A, Syed K, Sakhuja P, Malhotra V, Arora N, Sarin SK. Association of TNF-β polymorphism with disease severity among patients infected with hepatitis C virus. J Med Virol. 2004;72:60–65. doi: 10.1002/jmv.10533. [DOI] [PubMed] [Google Scholar]
- 4.Zecchina D, Novick M, Rubinstein V, Barak C, Dinarello H, Nagler A. Interleukin-18 binding protein in acute graft versus host disease and engraftment following allogeneic peripheral blood stem cell transplants. J Hematother Stem Cell Res. 2001;10(6):769–776. doi: 10.1089/152581601317210863. [DOI] [PubMed] [Google Scholar]
- 5.Kieszko R, Krawczyk P, Jankowska O, Chocholska S, Krol A, Milanowski J. The clinical significance of interleukin 18 assessment in sarcoidosis patients. Respir Med. 2006;101(4):722–728. doi: 10.1016/j.rmed.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Kaser D, Novick M, Rubeinstein B, Siegmund B, Enrich RO, Koch W, et al. Interferon-α induces interleukin-18 binding protein in chronic hepatitis C patients. Clin Exp Immunol. 2002;129(2):332–338. doi: 10.1046/j.1365-2249.2002.01911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asakawa M, Kono H, Amemiya H, Matsuda M, Suzuki T, Maki A, et al. Role of interleukin-18 and its receptor in hepatocellular carcinoma associated with hepatitis C virus infection. Int J Cancer. 2006;118(3):564–570. doi: 10.1002/ijc.21367. [DOI] [PubMed] [Google Scholar]
- 8.Yasuhiko K, Tomoaki H, Okamoto M, Kato S, Koda Y, Nagata N, et al. Enhanced expression of interleukin-18 and its receptor in idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. 2004;31(6):619–625. doi: 10.1165/rcmb.2003-0306OC. [DOI] [PubMed] [Google Scholar]
- 9.Zhang PA, Wu JM, Li Y, Yang XS. A general method for nested RT-PCR amplification and sequencing the complete HCV genotype 1 open reading frame. Virol J. 2005;2:88. doi: 10.1186/1743-422X-2-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura K, Kakimi K, Wieland S, Guidotti LG, Chisari FV. Interleukin-18 inhibits hepatitis B virus replication in the livers of transgenic mice. J Virol. 2002;76(21):10702–10707. doi: 10.1128/JVI.76.21.10702-10707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glas J, Torok HP, Tonenchi L, Kapser J, Schiemann U, Muller-Myhsok B, et al. Association of polymorphisms in the interleukin-18 gene in patients with Crohn’s disease depending on the CARD15/NOD2 genotype. Inflamm Bowel Dis. 2005;11(12):1031–1037. doi: 10.1097/01.MIB.0000187574.41290.b1. [DOI] [PubMed] [Google Scholar]
- 12.Tiret L, Godefroy T, Lubos E, Nicaud V, Tregouet DA, Barbaux S, et al. Genetic analysis of the interleukin-18 system highlights the role of the interleukin-18 gene in cardiovascular disease. Circulation. 2005;112(5):643–650. doi: 10.1161/CIRCULATIONAHA.104.519702. [DOI] [PubMed] [Google Scholar]
- 13.Segat L, Bevilacqua D, Boniotto M, Arraes LC, Souza PR, Lima Filho JL, et al. IL-18 gene promoter polymorphism is involved in HIV-1 infection in a Brazilian pediatric population. Immunogenetics. 2006;58(5–6):471–473. doi: 10.1007/s00251-006-0104-7. [DOI] [PubMed] [Google Scholar]
- 14.Goodman ZD, Ishak KG. Histopathology of hepatitis C virus infection. Semin Liver Dis. 1995;15(1):70–81, Review [DOI] [PubMed]
- 15.Garson JA, Tuke PW, Makris M, Briggs M, Machin SJ, Preston FE, et al. Demonstration of viraemia patterns in haemophiliacs treated with hepatitis-C-virus-contaminated factor VIII concentrates. Lancet. 1990;336(8722):1022–1025. doi: 10.1016/0140-6736(90)92487-3. [DOI] [PubMed] [Google Scholar]
- 16.Johns MB, Paulus-Thomas JE. Purification of human genomic DNA from whole blood using sodium perchlorate in place of phenol. Anal Biochem. 1989;180(2):276–278. doi: 10.1016/0003-2697(89)90430-2. [DOI] [PubMed] [Google Scholar]
- 17.Giedraitis V, He B, Huang WX, Hillert J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol. 2001;112:146–152. doi: 10.1016/S0165-5728(00)00407-0. [DOI] [PubMed] [Google Scholar]
- 18.Togbe D, Aurore G, Noulin N, Quesniaux VF, Schnyder-Candrian S, Schnyder B, et al. Nonredundant roles of TIRAP and MyD88 in airway response to endotoxin, independent of TRIF, IL-1 and IL-18 pathways. Lab Invest. 2006;86(11):1126–1135. doi: 10.1038/labinvest.3700473. [DOI] [PubMed] [Google Scholar]
- 19.Negishi H, Fujita Y, Yanai H, Sakaguchi S, Ouyang X, Shinohara M, et al. Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc Natl Acad Sci USA. 2006;103(41):15136–15141. doi: 10.1073/pnas.0607181103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abraham M, Shapiro S, Lahat N, Miller A. The role of IL-18 and IL-12 in the modulation of matrix metalloproteinases and their tissue inhibitors in monocytic cells. Int Immunol. 2002;14(12):1449–1457. doi: 10.1093/intimm/dxf108. [DOI] [PubMed] [Google Scholar]
- 21.Falasca K, Ucciferri C, Dalessandro M, Zingariello P, Mancino P, Petrarca C, et al. Cytokine patterns correlate with liver damage in patients with chronic hepatitis B and C. Ann Clin Lab Sci. 2006;36(2):144–150. [PubMed] [Google Scholar]
- 22.Murata K, Yamamoto N, Kawakita T, Saito Y, Yamanaka Y, Sugimoto K, et al. Up-regulation of IL-18 by interferon alpha-2b/ribavirin combination therapy induces an anti-viral effect in patients with chronic hepatitis C. Hepatogastroenterology. 2005;52(62):547–551. [PubMed] [Google Scholar]
- 23.Robertson MJ, Mier JW, Logan T, Atkins M, Koon H, Koch KM, et al. Clinical and biological effects of recombinant human interleukin-18 administered by intravenous infusion to patients with advanced cancer. Clin Cancer Res. 2006;12(14):4265–4273. doi: 10.1158/1078-0432.CCR-06-0121. [DOI] [PubMed] [Google Scholar]
- 24.Pawlik A, Kurzawski M, Czerny B, Gawronska-Szklarz B, Drozdzik M, Herczynska M. Interleukin-18 promoter polymorphism in patients with rheumatoid arthritis. Tissue Antigens. 2006;67(5):415–418 [DOI] [PubMed]
- 25.Mojtahedi Z, Naeimi S, Farjadian S, Omrani GR, Ghaderi A. Association of IL-18 promoter polymorphisms with predisposition to Type 1 diabetes. Diabet Med. 2006;23(3):235–239 [DOI] [PubMed]
- 26.Boraska V, Terzić J, Skrabić V, Caćev T, Bucević-Popović V, Peruzović M, et al. NeuroD1 gene and interleukin-18 gene polymorphisms in type 1 diabetes in Dalmatian population of Southern Croatia. Croat Med J. 2006;47(4):571–578 [PMC free article] [PubMed]
- 27.Lee HM, Park SA, Chung SW, Woo JS, Chae SW, Lee SH, et al. Interleukin-18/607 gene polymorphism in allegic rhinitis. Int J Pediatric Otorhinoalaryngol. 2006;70(6):1085–1088 [DOI] [PubMed]
- 28.Jang WC, Park SB, Nam YH, Lee SS, Kim JW, Chang IS, et al. Interleukin-18 gene polymorphisms in Korean patients with Behçet’s disease. Clin Exp Rheumatol. 2005;23(4 Suppl 38):S59–S63 [PubMed]
- 29.Zhou Y, Yamaguchi E, Fukui Y, Konno S, Maeda Y, Kimata K, et al. Enhanced expression of interleukin-18 receptor alpha chain by CD4+ T cells in sarcoidosis. Chest. 2005;128(4):2497–2503 [DOI] [PubMed]
- 30.Folwaczny M, Glas J, Török HP, Tonenchi L, Paschos E, Bauer B, et al. Polymorphisms of the interleukin-18 gene in periodontitis patients. J Clin Periodontol. 2005;32(5):530–534 [DOI] [PubMed]
- 31.Novak N, Kruse S, Potreck J, Maintz L, Jenneck C, Weidinger S, et al. Single nucleotide polymorphisms of IL-18 gene are associated with atopic eczema. J Allergy Clin Immunology. 2005;115(4):828–833 [DOI] [PubMed]
- 32.Aizawa Y, Sutoh S, Matsuoka M, Negishi M, Torii A, Miyakawa Y, et al. Association of interleukin-18 gene single nucleotide polymorphisms with susceptibility of inflammatory bowel disease. Tissue Antigens. 2005;65(1):88–92 [DOI] [PubMed]