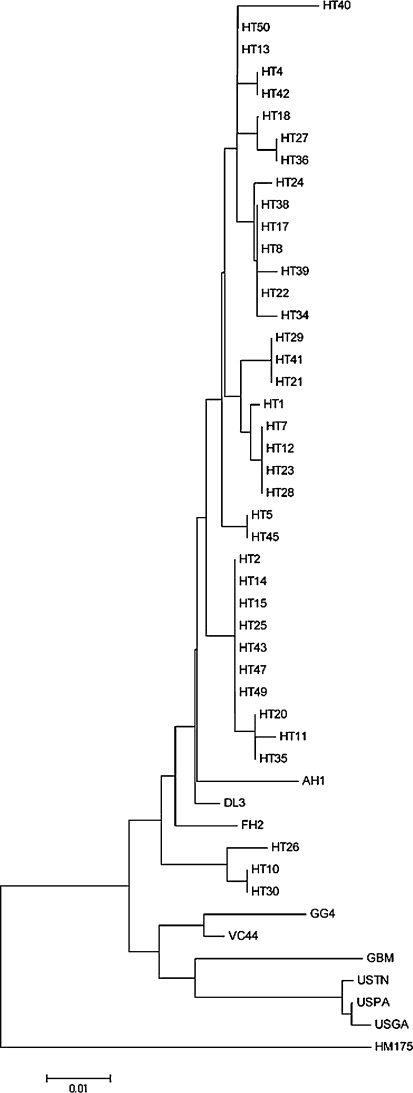
Fig. 4.
Phylogenetic analysis of the VP1/2A region of the strains isolated from Guigang and Hetian, and reported strains. Neighbor-joining phylogenetic trees of the VP1/2A region by using Kimura’s 2-parameter model. Strains are indicated at nodes. Bars denote genetic distances. HT indicates the strain isolated in Hetian, and GG4 refers to the strain isolated in Guigang. In Guigang, all 35 patients were infected by a single strain, and we selected GG4 to perform the phylogenetic analysis. AH1, AB020564 [15]; FH2, AB020568 [15]; VC44, AB253604; GBM, X75215 [16, 17]; HM175, M14707 [12], the DL3 sequence [11], and the strains isolated from the United States, USTN, USPA, and USGA