Abstract
Dopamine (DA) has been implicated as an endogenous neurotoxin to explain the selective neurodegeneration as observed for Parkinson's disease (PD). However, previous work demonstrated 3,4-dihydroxyphenylacetaldehyde (DOPAL) to be more toxic than DA. DOPAL is generated as a part of DA catabolism via the activity of monoamine oxidase and the mechanism of DOPAL toxicity is proposed to involve protein modification. Previous studies have demonstrated protein reactivity via the aldehyde moiety; however, DOPAL contains two reactive functional groups (catechol and aldehyde) both with the potential for protein adduction. The goal of this work was to determine whether protein modification by DOPAL occurs via a thiol-reactive quinone generated from oxidation of the catechol, which is known to occur for DA, or if the aldehyde forms adducts with amine nucleophiles. To accomplish this objective, the reactivity of DOPAL towards N-acetyl-lysine (NAL), N-acetyl-cysteine (NAC) and two model proteins was determined. In addition, several DOPAL analogues were obtained and used for comparison of reactivity. Results demonstrate that at pH 7.4 and 37°C, the order of DOPAL reactivity is NAL ≫ NAC and the product of NAL and DOPAL is stable in the absence of reducing agent. Moreover, DOPAL will react with model proteins, but in the presence of amine-selective modifiers citraconic anhydride and 2-iminothiolane hydrochloride, the reactivity of DOPAL towards the proteins is diminished. In addition, DOPAL-mediated protein cross-linking is observed when a model protein or a protein mixture (i.e. mitochondria lysate) are treated with DOPAL at concentrations of 5-100 μM. Protein cross-linking was diminished in the presence of ascorbate, suggesting the involvement of a quinone in DOPAL-mediated protein modification. These data indicate DOPAL to be highly reactive towards protein nucleophiles with the potential for protein cross-linking.
Introduction
Oxidative stress is hypothesized to play a significant role in Parkinson's disease (PD), a condition involving dopaminergic cell death (1, 2); however, the exact mechanisms underlying the pathogenesis of this common neurodegenerative disorder are currently unknown. It has been proposed that a toxin endogenous to dopamine neurons is generated subsequent to disease insult and this neurotoxin mediates neurodegeneration (3-6).
Studies have demonstrated that dopamine (DA) is an endogenous neurotoxin that readily undergoes auto-oxidation to an o-quinone (7, 8) capable of protein modification (9-11). Subsequent to biosynthesis, cytosolic DA is rapidly sequestered into vesicles and many reports have shown that dysregulation of DA trafficking and storage, such as toxicant-mediated release from vesicles, yields cell toxicity (12-14).
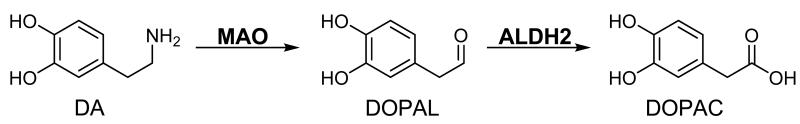
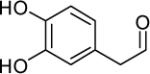
However, previous work has demonstrated that the aldehyde metabolite of DA, 3,4-dihydroxyphenylacetaldehyde (DOPAL), which results from the activity of monoamine oxidase (MAO) (Scheme 1), is orders of magnitude more toxic in vitro and in vivo than DA (3, 15). Physiologic concentrations of DOPAL are around 2-3 μM (16) and controlled via the action of aldehyde reductases and several aldehyde dehydrogenases, both cytosolic and mitochondrial (17). When levels of the aldehyde intermediate are only slightly elevated (6 μM), cell death is observed (18, 19). Such evidence indicates DOPAL to be toxic, and therefore, inhibition of its metabolism could be extremely harmful to cells and play a significant role in the onset of PD.
Scheme 1.
The primary pathway for DA metabolism involves oxidative deamination via MAO to an aldehyde intermediate followed by oxidation to an acid product.
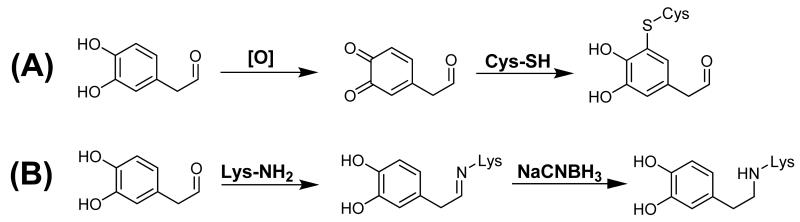
Several studies have demonstrated adduction of proteins by DOPAL (4, 20-22), and recent work has shown that inhibition of DOPAL oxidation and/or reduction via oxidative stress/lipid peroxidation products yielded an increase in [DOPAL] and protein modification (4, 23). However, protein residues susceptible to modification by DOPAL, i.e. Cys or Lys, have not been conclusively identified. Oxidation of the catechol yields a Cys-reactive quinone, which is known to occur for DA (7, 8, 24); however, the aldehyde may form adducts with Lys residues (4) (Scheme 2). In addition, the stability of each adduct under physiologic conditions is of question, especially for Lys as Schiff base products often require reduction for stability. It is also conceivable that DOPAL is a bifunctional electrophile which could cause extensive protein cross-linking. Therefore, the current study was undertaken to identify the mechanism by which DOPAL modifies proteins as the DA-derived aldehyde contains two reactive functional groups, i.e. aldehyde and catechol. DOPAL and structural analogues of DOPAL, including the DA metabolite 3-methoxy-4-hydroxyphenylacetaldehyde (MOPAL), were obtained and reacted with protein nucleophiles, including amino acids and two model proteins. For the first time, the protein reactivity of DOPAL was determined both qualitatively and quantitatively.
Scheme 2.
Possible mechanisms for DOPAL-mediated protein modification include (A) auto-oxidation of the catechol followed by reaction with protein thiols or (B) reaction with protein amines via the aldehyde. The Schiff base in (B) may require reduction for stability.
Experimental Procedures
Materials
DOPAL was biosynthesized via an established procedure involving enzyme-catalyzed conversion of DA to DOPAL by rat liver MAO (22) and its concentration was determined using an ALDH assay with nicotinamide adenine dinucleotide (NAD) (21) and high performance liquid chromatography (HPLC) analysis, as described below. 3,4-Dihydroxyphenylethanol (DOPET) was obtained via reduction of DOPAL with a 10-fold excess of sodium borohydride. The DOPAL quinone was generated via reaction of DOPAL with a 2-fold excess of sodium metaperiodate in 50 mM sodium phosphate buffer, pH 7.4 and used as a standard for HPLC analysis.
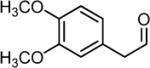
3,4-Dimethoxyphenylacetaldehyde (DMPAL) was synthesized via alkene oxidation of eugenol methyl ether (Sigma-Aldrich) using 3.5 mol% RuCl3 and 4 eq. NaIO4 [3 h in 6:1 acetonitrile:water; 40% yield; 1H NMR (CDCl3) δ 3.60 (d, 2H), 5.93 (s, 2H), 6.65-6.82 (m, 3H), 9.71 (t, 1H); 13C NMR (CDCl3) δ 50.23, 56.12 (2C), 111.82, 112.84, 122.03, 124.38, 148.55, 149.48, 199.68] (25).
MOPAL was biosynthesized via a similar procedure as outlined for DOPAL above, namely, rat liver MAO was used to convert 3-methoxytyramine to MOPAL.
DA, DOPAC, bovine serum album (BSA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from rabbit muscle and all other chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
Evaluation of the Reactivity of DOPAL
To determine the reactivity between DOPAL and Lys or Cys residues, 100 μM DOPAL was incubated with either N-acetyl-Lysine (NAL, 1, 2.5, 5, 7, 10 mM) or N-acetyl-Cysteine (NAC, 10 mM) (37°C, 50 mM sodium phosphate buffer, pH 7.4). Aliquots were taken periodically throughout the 90 min time course and the reaction was quenched by a 1:10 dilution with 1% trifluoroacetic acid (TFA) followed by storage at −20°C. The amount of DOPAL consumed during the course of the experiment was determined via HPLC as described below. In addition, a control was analyzed via HPLC to account for any spontaneous loss of DOPAL (i.e. oxidation to the acid, DOPAC).
Reactivity studies were also completed with reagents that were structurally similar to that of DOPAL. Specifically, NAL at varying concentrations was incubated for 3.5 h with 100 μM of either DMPAL, MOPAL or phenylacetaldehyde (PAL) in both the presence and absence of 5 mM sodium cyanoborohydride (NaCNBH3) reducing agent (50 mM sodium phosphate buffer, pH 7.4, 37°C). Aliquots were then quenched by a 1:10 dilution with 1% TFA and quantified by HPLC.
HPLC Analysis
Quantification of DMPAL, DOPAL, and PAL at each time point of the reactivity study was performed using an Agilent 1100 Series Capillary HPLC system with a photodiode array detector set to absorbance at 202 and 280 nm. Ten μL of sample solution was injected and separation was achieved using a Phenomenex C18 Luna microbore column (1 × 150 mm, 100 Å) and a mobile phase consisting of 0.1% TFA (v/v) in HPLC-grade water and 6% acetonitrile (v/v) at a flow rate of 50 μL/min. MOPAL was analyzed using a mobile phase consisting of 0.1% TFA (v/v) in HPLC-grade water and 10% acetonitrile (v/v) at a flow rate of 50 μL/min.
Comparison of Protein Reactivity, DOPAL vs. MOPAL
BSA (1 mg/mL) was treated with either 50 μM DOPAL, 50 μM MOPAL, 1:1 DOPAL:MOPAL (50 μM each), 1:2 DOPAL:MOPAL (50 μM and 100 μM) or 1:10 DOPAL:MOPAL (50 μM and 500 μM). In the presence of reducing agent (5 mM NaCNBH3), samples were incubated for 4 h (37°C, 50 mM sodium phosphate buffer, pH 7.4).
Catechol-containing protein adducts were detected using the redox-cycling dye nitroblue tetrazolium (NBT) (26, 27) or Coomassie Blue. While Coomassie stain detects all proteins, NBT is selective for catechols, which are redox-sensitive functional groups. Therefore, NBT will selectively stain proteins containing such adducts (26, 27). Samples (5 μg) were first subject to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 7.5% acrylamide) and the gels were placed in Bjenum and Schafer-Nielsen transfer buffer (37 °C, pH 7.4) for 10 min before being transferred to a nitrocellulose membrane utilizing a semi-dry transfer apparatus (70 min, 20 V). The nitrocellulose membrane was placed in 0.24 mM NBT with 2 M potassium glycine buffer (pH 10) and allowed to incubate overnight at 4 °C. The membranes were rinsed and stored with 5 mL distilled water (4 °C). The integrated density of each lane was quantified using the program NIH ImageJ v1.37 (http://rsb.info.nih.gov/ij/).
To further verify the compounds were binding to the protein during the course of the reaction, the aforementioned BSA samples treated with DOPAL and MOPAL were analyzed by HPLC using the procedure described above. In addition, a model peptide (10 μM RKRSRAE) was incubated with 100 μM of DOPAL or MOPAL (10 mM tricine buffer, pH 7.4, 4 h at 37 °C) and analyzed via MALDI-TOF mass spectrometry. Briefly, 1 μL of sample was diluted 1:1 with 0.1% TFA and mixed with 1 μL of water saturated with α-cyano-4-hydroxycinnaminic acid on a plate. The mixture was allowed to air dry and analyzed using a Bruker Biflex MALDI-TOF mass spectrometer in reflectron mode. Calibration was performed using the peptides angiotensin I and bradykinin.
Detection of DOPAL Adducts on Proteins Treated with a Lysine or Cysteine Modifier
The model proteins BSA (14 μM) and GAPDH (27.7 μM) were treated for 45 min at 37°C (50 mM sodium phosphate buffer, pH 7.4) with one of the following protein modifiers at concentrations of 0, 1, 5 and 10 mM; citraconic anhydride (modifies primary amines), 2-iminothiolane hydrochloride (Traut's reagent, converts amines to thiols (28)) or iodoacetic acid (modifies thiol residues). To verify the amine to thiol conversion, samples were titrated with 5,5′-dithiobis-(2-nitrobezoic acid) (DTNB) and absorbance at 412 nm was determined. Following reaction with the modifiers, the proteins were incubated with 100 μM DOPAL for 4 h at 37°C (50 mM sodium phosphate buffer, pH 7.4).
Additionally, BSA modified by citraconic anhydride (0, 1, 5, 10 mM) for 45 min at 37°C was treated with 1 M HCl to acidify the sample (pH 3) and incubated for 2 h to remove the citraconic anhydride adduct (Sigma Aldrich technical bulletin). It should be noted that citraconic anhydride can react with thiols; however, the resulting adduct cannot be removed via acid hydrolysis. Therefore, the ability to restore DOPAL-reactivity following acid hydrolysis of the citraconic anhydride modification allows discrimination between Lys and Cys adducts (29, 30). Samples were subsequently treated with 100 μM DOPAL and incubated overnight (37°C, 50 mM sodium phosphate buffer, pH 7.4).
As described above, DOPAL-protein adducts were analyzed by SDS-PAGE (7.5% or 10% acrylamide) followed by an overnight incubation with the NBT stain.
Determination of Catechol-Adducts on a Model Protein
To compare the reactivity of DA and its metabolites to adduct proteins, 100 μM DA, DOPAL, DOPAC, or 3,4-dihydroxyphenylalanine (L-DOPA) was incubated with 1 mg/mL (27.7 μM) GAPDH for either 3 or 6 h (37°C, 50 μM sodium phosphate buffer, pH 7.4). As described above, samples were analyzed via SDS-PAGE (10% acrylamide) followed by staining with NBT.
Presence of GAPDH Protein Cross-Linking upon Treatment with DOPAL
A 0.3 mg/mL solution of GAPDH (8.3 μM) was treated with DOPAL (0, 5, 50 and 100 μM) and incubated at 37°C for either 2 or 4 h (50 mM sodium phosphate buffer, pH 7.4). For comparison, the experiment was repeated using 0 and 100 μM MOPAL along with 0 and 100 μM DOPAL. Proteins were separated via SDS-PAGE (7.5% acrylamide) and stained with Coomassie Blue.
Additionally, a 0.3 mg/mL solution of GAPDH was treated with either DOPAL (0 and 50 μM), DOPAL (0 and 50 μM) with 5 mM NaCNBH3 or DOPAL (0 and 50 μM) with 5 mM sodium ascorbate and incubated at 37°C for 4 h (50 mM sodium phosphate buffer, pH 7.4). Samples were analyzed via SDS-PAGE (10% acrylamide) and stained with Coomassie Blue.
DOPAL-Protein Cross-Linking in a Protein Mixture
Mouse liver mitochondrial lysate was obtained from a 20-25 g Swiss Webster (CD1 IGS) mouse. Briefly, mice were euthanized via 150 mg/kg sodium pentobarbital administered I.P. and liver quickly removed. Tissue was homogenized in 10 mM tris buffer (pH 7.4) containing 0.25 M sucrose solution at 4 °C, and samples were centrifuged at 1000 g for 10 min. The supernatant was collected and spun at 12,000 g for 10 min, and the pellet was reconstituted in homogenizing media (4 °C). This step was repeated for a total of three spins, yielding mitochondria. The mitochondria were reconstituted in 50 mM sodium phosphate buffer (pH 7.4) lysed via sonication and membranes pelleted via centrifugation at 100,000 g for 30 min (4 °C). Mouse liver mitochondria lysate (0.5 mg/mL) was incubated with DOPAL (0 and 10 μM) at 37°C for 2, 4 or 6 h (50 mM sodium phosphate buffer, pH 7.4). To samples containing an initial DOPAL concentration of 10 μM, additional DOPAL was added every 1 h (5 μM final). Samples were analyzed via SDS-PAGE (7.5% acrylamide) and stained with Coomassie Blue. To determine the extent of DOPAL metabolism during the course of the reaction, the 6 h sample was analyzed by HPLC after treatment with perchloric acid (5% v/v) to precipitate protein.
Results
DOPAL is more reactive towards Lys than Cys residues
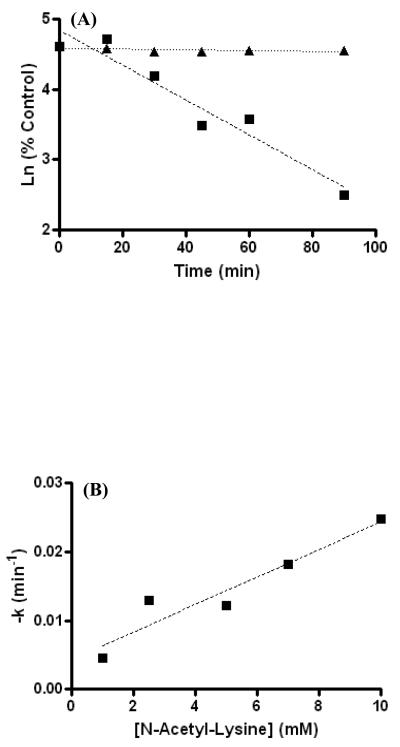
To determine the ability of DOPAL to modify amine vs thiol nucleophiles, the reactivity of DOPAL towards NAL and NAC was monitored using HPLC over a 1.5 h time course. As demonstrated by data in Figure 1A, no observable reaction occurred between DOPAL and NAC under conditions used; however, DOPAL showed considerable reactivity towards NAL. Slopes (−k′) for decreasing [DOPAL] were log linear and correlated with [NAL]. The rate constant for the DOPAL-NAL reaction was calculated to be 2.0 M−1min−1 (Figure 1B).
Figure 1.
(A) No apparent reaction occurs between DOPAL (100 μM) and NAC (▲) (10 mM) over the indicated time course whereas a considerable reaction appears to occur between DOPAL (100 μM) and NAL (■) (10 mM). Such a result indicates that DOPAL is more reactive with amine nucleophiles than thiol nucleophiles. (B) Treatment of NAL at various concentrations with 100 μM DOPAL yields a second order rate constant of 2.0 M−1min−1.
Both the catechol and aldehyde functional groups are required for protein modification
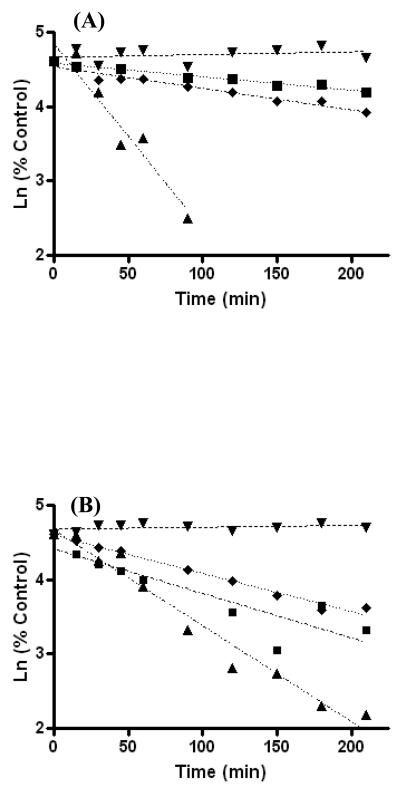
Compounds structurally analogous to DOPAL were obtained and utilized to study the reaction between DOPAL and NAL (Figure 2). These analogues either lack a catechol or contain a protected catechol. In the absence of reducing agent (Figure 2A), DOPAL was observed to be 5-fold more reactive towards NAL than MOPAL and 10-fold more reactive than PAL. In the presence of NaCNBH3, the reactivity of MOPAL and PAL towards NAL is increased however, it is still significantly less than the reaction between DOPAL and NAL reaction (Figure 2B). Such a finding indicates DOPAL to be highly reactive towards amine nucleophiles, more so than DMPAL, MOPAL or PAL. In addition, DOPAL will form a stable adduct such that reduction to an amine is not necessary, suggesting the formation of a reversible Schiff base-type adduct.
Figure 2.
Reaction between NAL and DOPAL (▲) and NAL with various structural analogous of DOPAL (DMPAL (▼), MOPAL (◆) and PAL (■)), indicate a catechol is essential for enhanced reactivity with amine nucleophiles. In the (A) absence and (B) presence of 5 mM NaCNBH3, DOPAL is highly reactive towards NAL and will form a stable adduct such that reduction to an amine is not necessary. The results demonstrate the order of reactivity towards NAL to be DOPAL ≫ PAL, MOPAL > DMPAL and are further summarized in Table 1.
Additionally, a comparison of the extent of protein modification by DOPAL versus MOPAL (Figure 3A) further demonstrates the greater reactivity of DOPAL than MOPAL and it's ability to form stable protein adducts. In the presence of increased MOPAL concentration (i.e. 1:2 and 1:10 ratios), the % of NBT staining as compared to the control decreased only slightly indicating that there was little competition for protein amines between DOPAL and MOPAL. If MOPAL was competing for Lys residues, NBT staining would be significantly diminished as NBT detects the presence of catechols and MOPAL's structure does not contain a catechol. To further study the competition of DOPAL and MOPAL for protein nucleophiles, the samples were analyzed via HPLC to measure changes in [DOPAL] and [MOPAL]. It was determined that MOPAL did not positively or negatively influence DOPAL adduction during the course of the reaction.
Figure 3.
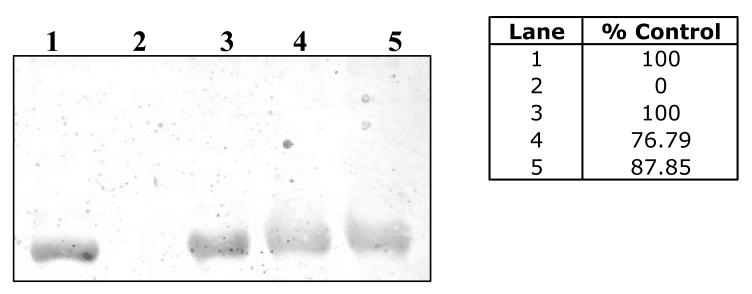
DOPAL-mediated protein modification is not hindered in the presence of MOPAL. Treatment of BSA with DOPAL (lane 1), MOPAL (lane 2), 1:1 DOPAL:MOPAL (lane 3), 1:2 DOPAL: MOPAL (lane 4) and 1:10 DOPAL:MOPAL (lane 5). % Control refers to the integrated staining density for the control sample (BSA + DOPAL).
To confirm protein/amine reactivity of MOPAL, a model peptide was incubated with MOPAL and analyzed via MALDI-TOF mass spectrometry. Treatment of RKRSRAE (measured m/z at 901.974) with MOPAL yielded new peaks corresponding to the peptide with 150.00 Da adducts (i.e. m/z at 1051.946, 1201.946). The shift in mass of 150.00 Da corresponds to a MOPAL adduct generated via reaction of MOPAL with Lys followed by reduction of the imine to an amine (theoretical mass for adduct = 150.06 Da).
Treatment of DOPAL with the peptide RKRSRAE yielded a new peak at m/z 1037.7 corresponding to a 134.3 Da adduct and Schiff base formation as previously described (4).
Reactivity of DOPAL towards proteins is diminished when proteins are pre-treated with amine modifiers
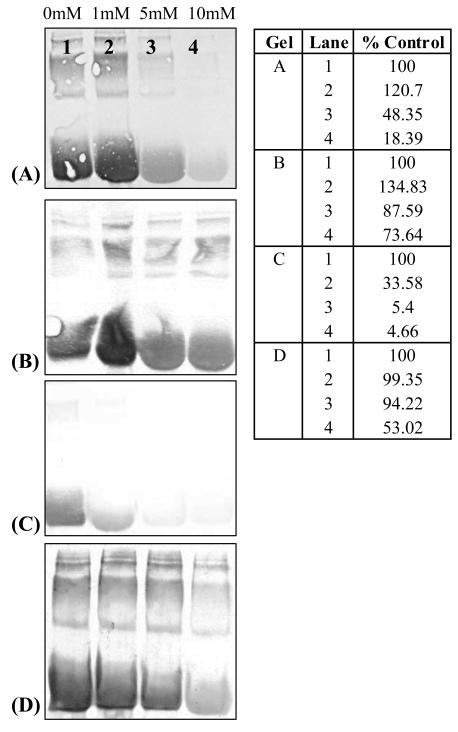
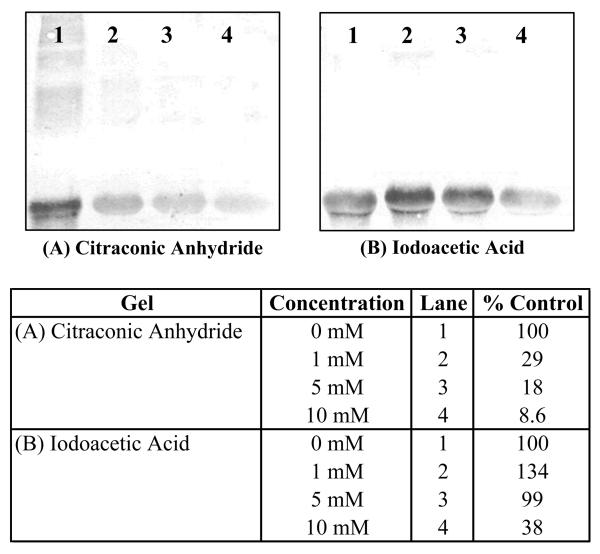
As shown in Figures 4 and 5, pre-treatment of BSA and GAPDH with the protein modifiers citraconic anhydride (modifies primary amines), Traut's reagent (converts amines to thiols (28)) and iodoacetic acid (modifies thiol residues) further demonstrates the reactivity of DOPAL towards amine nucleophiles. Treatment of BSA with increasing concentrations of citraconic anhydride reduced the number of adducts on BSA, as demonstrated in Figure 4A. Upon removal of the citraconic anhydride adduct, DOPAL reactivity was restored (Figure 4B) thus further indicating DOPAL protein modification is dependent upon the availability of amine residues. When BSA was pre-treated with Traut's reagent, DOPAL-protein adducts were significantly diminished (Figure 4C), whereas, pre-treatment with iodoacetic acid, did not significantly affect DOPAL's ability to modify the protein (Figure 4D). To verify the effect of Traut's reagent on the protein, samples were titrated with DTNB and an increase in free thiol concentration was observed, indicating an amine to thiol conversion (data not shown).
Figure 4.
The reactivity of DOPAL towards BSA is diminished when the model protein is pre-treated with (A) citraconic anhydride (modifies amines) however; reactivity of DOPAL is restored when the (B) citraconic anhydride moiety is removed. Additionally, DOPAL reactivity is reduced when BSA is pre-treated with (C) Traut's reagent (converts amines to thiols) but is not affected when BSA is pre-treated with (D) iodoacetic acid (modifies thiols). These results indicate that DOPAL modifies amine residues. For all lanes, the concentration of the protein modifiers are 0 mM (lane 1), 1 mM (lane 2), 5 mM (lane 3) and 10 mM (lane 4). % Control refers to the integrated staining density for the control sample (untreated BSA).
Figure 5.
The reactivity of DOPAL towards GAPDH is diminished when the protein is pre-treated with (A) Traut's reagent (converts amines to thiols) however; when GAPDH is pre-treated with (B) iodoacetic acid (modifies thiols), protein modification via DOPAL is not affected. For all lanes, the concentration of the protein modifiers are 0 mM (lane 1), 1 mM (lane 2), 5 mM (lane 3) and 10 mM (lane 4). % Control refers to the integrated staining density relative to that of the control sample (untreated GAPDH).
In line with such findings, data in Figure 5A demonstrate that pre-treatment of GAPDH with Traut's reagent diminished the reactivity of DOPAL towards the protein. When GAPDH was pre-treated with iodoacetic acid, protein modification via DOPAL was not significantly affected unless high concentration (i.e. 10 mM) of iodoacetic acid was used (Figure 5B).
Modification of GAPDH by DA and its metabolites
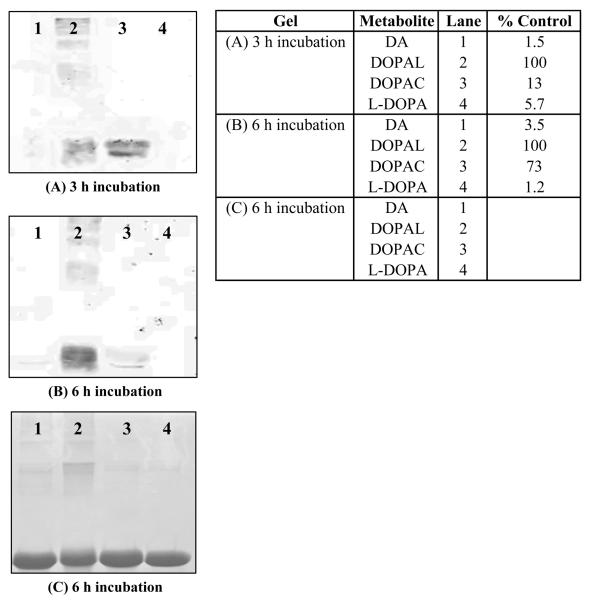
As shown in Figure 6A,B, treatment of GAPDH with DA, DOPAL, DOPAC or L-DOPA (100 μM) resulted in varying levels of NBT reactivity and protein modification. The order of reactivity, based on NBT staining, was determined to be DOPAL > DOPAC > DA, L-DOPA. Additionally, these results indicate that DOPAL (Lane 2) crosslinks GAPDH as demonstrated by the presence of higher molecular weight protein bands, indicative of GAPDH oligomers such as the tetramer (i.e. 146 kD tetramer).
Figure 6.
GAPDH incubated with 100 μM of DA, DOPAL, DOPAC or L-DOPA for (A) 3 h and (B) 6 h yielded varying degrees of reactivity towards the model protein. Results demonstrate the order of reactivity to be DOPAL > DOPAC > L-DOPA, DA. In addition, protein cross-linking of GAPDH by DOPAL was observed using two staining methods: (A), (B) NBT and (C) Coomassie. % Max signal refers to the integrated staining density for a given lane relative to that determined for lane 2 (GAPDH + DOPAL), which had the highest staining intensity.
DOPAL mediates GAPDH protein cross-linking
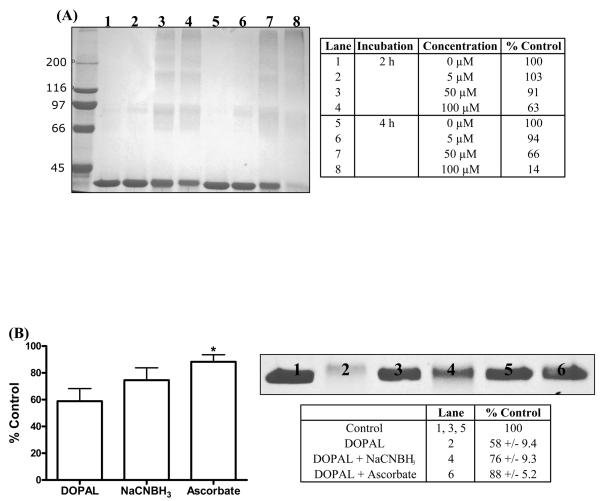
Protein cross-linking was evident for GAPDH treated with DOPAL at various concentrations. As shown in Figure 7A, control samples at 2 and 4 h contain a protein band at 36 kDa, corresponding to the molecular weight of the GAPDH monomer. However, in samples treated with DOPAL (5-50 μM), higher molecular weight protein bands became apparent. Specifically, protein treated with 100 μM DOPAL yielded protein bands in excess of 200 kDa while the band corresponding to the 36 kDa monomer is reduced in staining intensity at both 2 and 4 h (37 and 86%, respectively).
Figure 7.
(A) Treatment of GAPDH with various concentrations of DOPAL (5, 50 and 100 μM) demonstrate the ability for DOPAL to cause protein cross-linking of GAPDH monomers. % Control refers to the integrated staining density for the GAPDH monomer (37 kD) relative to that of the control samples (untreated protein). (B) The presence of 5 mM NaCNBH3 (lane 4) and 5 mM ascorbate (lane 6) will protect GAPDH against DOPAL-induced protein cross-linking. % Control refers to the integrated staining density relative to that of the control samples (untreated GAPDH). The band shown represents the 37 kD GAPDH protein. Values shown represent means ± SEM (n = 4). *, significantly different from GAPDH treated with DOPAL (p < 0.05), determined using a two-tailed t-test.
When GAPDH is treated with DOPAL in the presence of NaCNBH3 (imine and quinone reducing agent) and ascorbate (quinone reducing agent), protein cross-linking/aggregation was significantly diminished (Figure 7B). Reducing agents appear to offer protection from protein cross-linking via DOPAL, with ascorbate being the most effective.
Interestingly, GAPDH treated with 100 μM MOPAL did not yield any noticeable protein cross-linking, in contrast to that observed for GAPDH incubated with DOPAL (data not shown).
DOPAL-mediated protein cross-linking in protein mixture
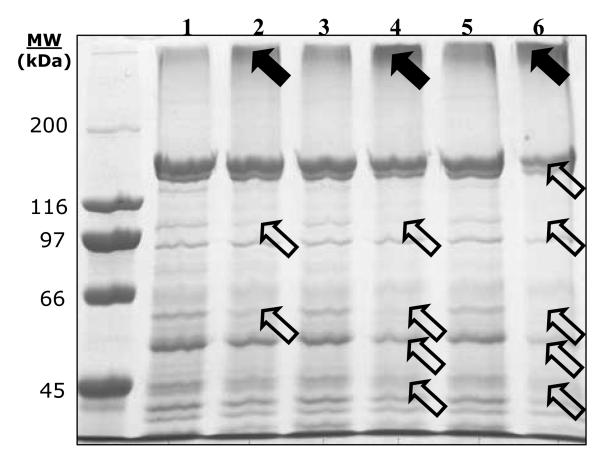
When a mixture of proteins (i.e. mouse liver mitochondrial lysate) was treated with DOPAL, protein modification was observed at all three time points; 2, 4 and 6 h (Figure 8). High molecular weight protein bands (i.e. in excess of 200 kDa) increased in staining, while proteins at lower molecular weights (i.e. 57, 67 and 90 kDa) decreased in signal as compared to controls. Such evidence indicates the ability for DOPAL to cause protein cross-linking. Analysis by HPLC found the concentration of DOPAL in the 6 h sample to be 12 μM.
Figure 8.
Lysate from mouse liver mitochondria (0.5 mg/mL) treated with DOPAL resulted in time-dependent protein cross-linking. Lanes 1, 3 and 5 represent control protein incubated without DOPAL for 2, 4 and 6 h, respectively. Lanes 2, 4 and 6 represent protein treated with DOPAL for 2, 4 and 6 h as described in the Material and Methods section. Solid and hollow arrows denote an increase and decrease in protein band intensity, respectively.
Discussion
The DA intermediate, DOPAL, has been implicated as an endogenous neurotoxin relevant to the pathogenesis of PD (8, 11). Studies have demonstrated DOPAL to be toxic towards dopaminergic cells via various mechanisms including production of oxidative stress, induction of the mitochondrial transition pore and protein modification (3, 15, 31, 32). Recent work has shown that products of lipid peroxidation (i.e., 4-hydroxynonenal and malondialdehyde) will inhibit DA catabolism, specifically, the aldehyde oxidation step catalyzed by one or more aldehyde dehydrogenases. Such inhibition leads to increased concentrations of DOPAL using rat brain mitochondria and striatal synaptosomes as model systems (4, 23). Evidence of protein modification via DOPAL was demonstrated including indication that DOPAL has a significantly greater reactivity towards proteins than DA or DOPAC (4).
The current study further extends these previous findings with the goal of identifying the mechanism by which DOPAL modifies protein. Data are presented demonstrating the reactivity between DOPAL and Lys is greater than that for the reactivity between DOPAL and Cys; however, protein cross-linking, mediated by a DOPAL-quinone, was observed in experiments utilizing model proteins and mouse liver mitochondria.
DOPAL is predicted to be a bifunctional electrophile as it contains two reactive functional groups (i.e. aldehyde and catechol). Therefore, it was of interest to determine if DOPAL-mediated protein modification occurs by oxidation of the catechol to yield a Cys-reactive quinone (which is known to occur for DA (7, 8)) or if the aldehyde forms adducts with Lys residues. As demonstrated by data in Figure 1, the reaction of DOPAL with NAL was found to be significant, as demonstrated by the time-dependent loss of [DOPAL]. However, there was no significant reaction observed between DOPAL and NAC under the conditions used. Such a finding indicates that amine adduction may represent a significant pathway for DOPAL-protein modification. Although ortho-quinones are highly reactive toward protein thiols, it should be noted that oxidation/activation of the catechol is first required and is rapidly followed by rearrangement or intracyclization (7, 8, 24). In contrast, the aldehyde moiety does not require activation or oxidation before protein modification. During the course of our reactions, DOPAL was not oxidized to an o-quinone as determined using HPLC and a DOPAL o-quinone standard generated via tyrosinase or sodium metaperiodate. However, under significant oxidative conditions, such as that found in the substantia nigra during DA catabolism (i.e. hydrogen peroxide production), or perhaps in the presence of prostaglandin-H-synthase, significant catechol oxidation could occur to a greater extent (33, 34).
While the aldehyde appears to be more reactive than the catechol toward protein nucleophiles under the conditions used, modification of the catechol greatly alters DOPAL reactivity, indicating that the ability of DOPAL to react with amine nucleophiles is dependent on the catechol (Figure 2 and Table 1). The structurally analogous compounds MOPAL, DMPAL and PAL were incubated with NAL, and it was found that DOPAL is much more reactive toward NAL, even in the presence of NaCNBH3. It is apparent from these results that the reactivity of these compounds towards NAL is dependent upon the structural differences of the catechol moiety, i.e. aryl substituents. This raises the question: how do the aryl substituents enhance the reaction rate? It is conceivable that the observed effect of catechol substitutions is due to 1). Increased aldehyde reactivity, or 2). Increased stability of the product.
Table 1.
Comparison of reactivity for DOPAL and analogues.
Reducing agent (NaCNBH3) included for stability. Without reducing agent, reactivity was very low, << 0.40 M−1min−1
None Detected. No significant reaction detected during the time-course.
Very low reactivity, estimated to be < 0.2 M−1min−1
Interestingly, the reactivity of MOPAL and PAL towards NAL increases in the presence of NaCNBH3, while the reactivity of DOPAL significantly decreases in the presence of reducing agent (Figure 2). NaCNBH3 is an imine- and quinone-reducing agent, and therefore, the enhanced reactivity of MOPAL/PAL in the presence of NaCNBH3 is predicted to stem from Schiff base (unstable) reduction to an amine (stable). In contrast, the DOPAL adduct could oxidize to a quinone that rearranges to a more stable form (e.g. indole), via reaction of the amine with the quinone. Therefore, a reducing agent is expected to preclude oxidation of the catechol and subsequent rearrangement to a stable adduct. In support of this assertion, it was found that inclusion of 5 mM ascorbate in incubations of DOPAL with NAL also significantly decreased the apparent rate of the reaction (data not shown). Such evidence implicates a role for a quinone in DOPAL-protein modification and indicates that the reaction of DOPAL with protein amines is dependent on catechol oxidation. Work is in progress to chemically characterize the structure of DOPAL-amino acid conjugates.
To investigate further the reactivity of DOPAL and the structurally analogous compound, MOPAL, with protein nucleophiles, BSA was treated with varying ratios of MOPAL and DOPAL. It should be noted that MOPAL is an endogenous product of DA catabolism and is found within dopaminergic cells via activity of catechol-O-methyl transferase with DOPAL and from MAO with 3-methoxytyramine (35). Due to the simultaneous generation of these two compounds in the cell, it is predicted that MOPAL competes for protein binding sites with DOPAL. While the reaction rate for DOPAL with NAL was found to be much higher than that for MOPAL (Figure 2, Table 1), an additional experiment was performed to show conclusively that DOPAL is more reactive than MOPAL. Using a model peptide containing primary amines, it was found that indeed, MOPAL readily reacts with Lys to form a Schiff-base product. The data in Figure 3, however, demonstrate that MOPAL does not significantly interfere with DOPAL-protein adduction. Specifically, DOPAL is able to modify protein to a similar extent in both absence and excess of MOPAL (i.e. 1:10 ratio). In addition, these findings suggest that methylation of DOPAL catechol-O-methyl transferase may be a detoxication step.
Use of NAL and NAC as models for protein nucleophiles provided evidence that amines are reactive toward DOPAL; however, it was of interest to determine the extent to which amines are responsible for DOPAL adducts on proteins. Two model proteins were utilized for this work, BSA and GAPDH. As shown in Figures 4 and 5, when primary amines were selectively modified (citraconic anhydride) and/or converted to thiol residues (Traut's reagent), the reactivity of DOPAL towards BSA or GAPDH was significantly diminished, as judged via NBT staining. Removal of the citraconic anhydride adduct (via acid) restored protein reactivity toward DOPAL, indicating involvement of Lys. In addition, if Cys were the primary target of DOPAL, then conversion of Lys to Cys via the Traut's reagent would yield an increase in DOPAL-protein modification. In contrast, when thiol residues were selectively modified (i.e. iodoacetic acid) no significant change in protein modification was observed except at high [iodoacetic acid] (10 mM). These results further demonstrate that reaction of DOPAL with proteins involves Lys/primary amines to a significant extent.
In a previous study, it was determined that DOPAL is more reactive towards BSA than DA or DOPAC (4). In line with such findings, the data in Figure 6 demonstrate a similar result for the model protein GAPDH. Specifically, DOPAL was found to be more reactive towards GAPDH than DOPAC, DA or L-DOPA. In addition, treatment of protein with DOPAL for 3 or 6 h, resulted in higher molecular weight protein bands corresponding to various oligomers and aggregates of GAPDH that were resistant to the reducing and denaturing conditions used in SDS-PAGE. This result was observed only for DOPAL and not the other related compounds (i.e., LDOPA, DA, DOPAC). It should be noted that such a finding is similar to that reported by Burke et al., who demonstrated that incubation of DOPAL with α-synuclein resulted in significant protein aggregation (36).
In vivo, GAPDH can exist as a tetramer containing 37 kD subunits, as a dimer or as the 37 kDa monomer, depending on the specific function that is needed (37). In the presence of DOPAL, significant cross-linking between GAPDH subunits was demonstrated (Figure 7A). This result was not observed for DA, DOPAC, L-DOPA or MOPAL, with the latter indicating that protein aggregation is dependent on the catechol. Evidence of protein cross-linking becomes more apparent with increasing DOPAL concentration and length of incubation time.
Of question then, is how is DOPAL cross-linking GAPDH? As noted above, while MOPAL is protein reactive, treatment of GAPDH with the DOPAL analogue (i.e. protected catechol) did not result in protein aggregation (data not shown). Therefore, it was hypothesized that DOPAL-mediated cross-linking occurs via Lys modification followed by catechol oxidation and Cys adduction. To test this hypothesis, GAPDH was treated with both DOPAL and the quinone-reducing agent, ascorbate (38). It was found that the presence of ascorbate afforded protection against DOPAL-mediated protein aggregation as shown in Figure 7B, and such a result indicates the involvement of a quinone in DOPAL-mediated protein cross-linking. Interestingly, NaCNBH3 also protected against protein cross-linking, which is different than predicted given that the reducing agent will stabilize the DOPAL adduct (i.e. reduce imine to amine); however, NaCNBH3 can also reduce quinones (39). These results implicate the possible involvement of catechol oxidation in the mechanism of DOPAL-mediated protein modification and cross-linking.
To determine the extent of protein cross-linking in a complex mixture of proteins, mitochondrial lysate was obtained and incubated with DOPAL. As shown in Figure 8, a shift from lower to higher molecular weight bands was observed as compared to control. Specific proteins decreased in staining intensity over time in the presence of DOPAL, and in particular, a large band appeared at approximately the 200 kDa molecular weight marker.
An increase in DOPAL concentration could therefore be detrimental to cells in regards to protein modification/cross-linking. Physiologic concentrations of DOPAL are estimated to be around 2-3 μM (16) and several studies have demonstrated that the DA-derived aldehyde at concentrations as low as 6 μM, just above basal level, can cause cell death over time (10, 11). The data in Figure 8, demonstrate a potential consequence of DOPAL at low μM in vitro.
In summary, the present study qualitatively and quantitatively demonstrated the reactivity of DOPAL towards protein nucleophiles. Specifically, it was determined that DOPAL is significantly more reactive towards Lys than Cys. Such evidence demonstrates aldehyde reactivity, however, the catechol greatly influences Lys modification. In addition, using model proteins (BSA, GAPDH) and a protein mixture (mitochondria lysate) treated with DOPAL (5-100 μM), considerable protein cross-linking was demonstrated. These data demonstrate DOPAL to be a bifunctional electrophile involving both the aldehyde and catechol in its mechanism of protein modification. These results may prove useful for predicting target proteins and sites of adduction on proteins.
Acknowledgements
This work was supported by grants NIH R01 ES15507 (J.A.D.), NIH K22 ES12982 (J.A.D.), a pilot grant via NIH P30 ES05605 (J.A.D.), and a fellowship via NIH T32 GM067795 (L.L.E.).
Abbreviations
- DTNB
5,5′-dithiobis-(2-nitrobezoic acid)
- NAC
N-acetyl-cysteine
- NAL
N-acetyl-lysine
- ALDH
aldehyde dehydrogenase
- AR
aldehyde or aldose reductase
- BSA
bovine serum albumin
- HPLC
high performance liquid chromatography
- L-DOPA
3,4-dihydroxyphenylalanine
- DA
dopamine
- DMPAL
3,4-dimethoxyphenylacetaldehyde
- DOPAC
3,4-dihydroxyphenylacetic acid
- DOPAL
3,4-dihydroxyphenylacetaldehyde
- DOPET
3,4-dihydroxyphenylethanol
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- MALDI-TOFMS
matrix-assisted laser desorption/ionzation-time of flight mass spectrometry
- MAO
monoamine oxidase
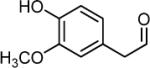
- MOPAL
3-methoxy-4-hydroxyphenylacetaldehyde
- NaCNBH3
sodium cyanoborohydride
- NAD
nicotinamide adenine dinucleotide
- NBT
nitroblue tetrazolium
- PD
Parkinson's disease
- PAL
phenylacetaldehyde
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TFA
trifluoroacetic acid
References
- 1.Jenner P. Oxidative stress in Parkinson's disease. Ann Neurol. 2003;53(Suppl 3):S26–36. doi: 10.1002/ana.10483. discussion S36-28. [DOI] [PubMed] [Google Scholar]
- 2.Zhou C, Huang Y, Przedborski S. Oxidative stress in Parkinson's disease: a mechanism of pathogenic and therapeutic significance. Ann N Y Acad Sci. 2008;1147:93–104. doi: 10.1196/annals.1427.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke WJ, Li SW, Williams EA, Nonneman R, Zahm DS. 3,4-Dihydroxyphenylacetaldehyde is the toxic dopamine metabolite in vivo: implications for Parkinson's disease pathogenesis. Brain Res. 2003;989:205–213. doi: 10.1016/s0006-8993(03)03354-7. [DOI] [PubMed] [Google Scholar]
- 4.Rees JN, Florang VR, Anderson DG, Doorn JA. Lipid peroxidation products inhibit dopamine catabolism yielding aberrant levels of a reactive intermediate. Chem Res Toxicol. 2007;20:1536–1542. doi: 10.1021/tx700248y. [DOI] [PubMed] [Google Scholar]
- 5.Collins MA, Neafsey EJ. Potential neurotoxic “agents provocateurs” in Parkinson's disease. Neurotoxicol Teratol. 2002;24:571–577. doi: 10.1016/s0892-0362(02)00210-6. [DOI] [PubMed] [Google Scholar]
- 6.Ramsden DB, Parsons RB, Ho SL, Waring RH. The aetiology of idiopathic Parkinson's disease. Mol Pathol. 2001;54:369–380. [PMC free article] [PubMed] [Google Scholar]
- 7.Graham DG. Oxidative pathways for catecholamines in the genesis of neuromelanin and cytotoxic quinones. Mol Pharmacol. 1978;l4:633–643. [PubMed] [Google Scholar]
- 8.Graham DG, Tiffany SM, Bell WR, Jr., Gutknecht WF. Autoxidation versus covalent binding of quinones as the mechanism of toxicity of dopamine, 6-hydroxydopamine, and related compounds toward C1300 neuroblastoma cells in vitro. Mol Pharmacol. 1978;14:644–653. [PubMed] [Google Scholar]
- 9.Conway KA, Rochet JC, Bieganski RM, Lansbury PT., Jr. Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science. 2001;294:1346–1349. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- 10.Hastings TG, Lewis DA, Zigmond MJ. Reactive dopamine metabolites and neurotoxicity: implications for Parkinson's disease. Adv Exp Med Biol. 1996;387:97–106. doi: 10.1007/978-1-4757-9480-9_13. [DOI] [PubMed] [Google Scholar]
- 11.LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Dopamine covalently modifies and functionally inactivates parkin. Nat Med. 2005;11:1214–1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- 12.Kitazawa M, Anantharam V, Kanthasamy AG. Dieldrin-induced oxidative stress and neurochemical changes contribute to apoptopic cell death in dopaminergic cells. Free Radic Biol Med. 2001;31:1473–1485. doi: 10.1016/s0891-5849(01)00726-2. [DOI] [PubMed] [Google Scholar]
- 13.Caudle WM, Colebrooke RE, Emson PC, Miller GW. Altered vesicular dopamine storage in Parkinson's disease: a premature demise. Trends Neurosci. 2008;31:303–308. doi: 10.1016/j.tins.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Ding Y, Cagniard B, Van Laar AD, Mortimer A, Chi W, Hastings TG, Kang UJ, Zhuang X. Unregulated cytosolic dopamine causes neurodegeneration associated with oxidative stress in mice. J Neurosci. 2008;28:425–433. doi: 10.1523/JNEUROSCI.3602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke WJ. 3,4-dihydroxyphenylacetaldehyde: a potential target for neuroprotective therapy in Parkinson's disease. Curr Drug Targets CNS Neurol Disord. 2003;2:143–148. doi: 10.2174/1568007033482913. [DOI] [PubMed] [Google Scholar]
- 16.Burke WJ, Chung HD, Li SW. Quantitation of 3,4-dihydroxyphenylacetaldehyde and 3, 4-dihydroxyphenylglycolaldehyde, the monoamine oxidase metabolites of dopamine and noradrenaline, in human tissues by microcolumn high-performance liquid chromatography. Anal Biochem. 1999;273:111–116. doi: 10.1006/abio.1999.4196. [DOI] [PubMed] [Google Scholar]
- 17.Marchitti SA, Deitrich RA, Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol Rev. 2007;59:125–150. doi: 10.1124/pr.59.2.1. [DOI] [PubMed] [Google Scholar]
- 18.Kristal BS, Conway AD, Brown AM, Jain JC, Ulluci PA, Li SW, Burke WJ. Selective dopaminergic vulnerability: 3,4-dihydroxyphenylacetaldehyde targets mitochondria. Free Radic Biol Med. 2001;30:924–931. doi: 10.1016/s0891-5849(01)00484-1. [DOI] [PubMed] [Google Scholar]
- 19.Mattammal MB, Haring JH, Chung HD, Raghu G, Strong R. An endogenous dopaminergic neurotoxin: implication for Parkinson's disease. Neurodegeneration. 1995;4:271–281. doi: 10.1016/1055-8330(95)90016-0. [DOI] [PubMed] [Google Scholar]
- 20.Helander A, Tottmar O. Reactions of biogenic aldehydes with hemoglobin. Alcohol. 1989;6:71–75. doi: 10.1016/0741-8329(89)90076-1. [DOI] [PubMed] [Google Scholar]
- 21.Ungar F, Tabakoff B, Alivisatos SG. Inhibition of binding of aldehydes of biogenic amines in tissues. Biochem Pharmacol. 1973;22:1905–1913. doi: 10.1016/0006-2952(73)90050-6. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson GE, Tottmar O. Biogenic aldehydes in brain: on their preparation and reactions with rat brain tissue. J Neurochem. 1987;48:1566–1572. doi: 10.1111/j.1471-4159.1987.tb05702.x. [DOI] [PubMed] [Google Scholar]
- 23.Florang VR, Rees JN, Brogden NK, Anderson DG, Hurley TD, Doorn JA. Inhibition of the oxidative metabolism of 3,4-dihydroxyphenylacetaldehyde, a reactive intermediate of dopamine metabolism, by 4-hydroxy-2-nonenal. Neurotoxicology. 2007;28:76–82. doi: 10.1016/j.neuro.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Tse DC, McCreery RL, Adams RN. Potential oxidative pathways of brain catecholamines. J Med Chem. 1976;19:37–40. doi: 10.1021/jm00223a008. [DOI] [PubMed] [Google Scholar]
- 25.Yang D, Zhang C. Ruthenium-catalyzed oxidative cleavage of olefins to aldehydes. J Org Chem. 2001;66:4814–4818. doi: 10.1021/jo010122p. [DOI] [PubMed] [Google Scholar]
- 26.Paz MA, Fluckiger R, Boak A, Kagan HM, Gallop PM. Specific detection of quinoproteins by redox-cycling staining. J Biol Chem. 1991;266:689–692. [PubMed] [Google Scholar]
- 27.Akagawa M, Ishii Y, Ishii T, Shibata T, Yotsu-Yamashita M, Suyama K, Uchida K. Metal-catalyzed oxidation of protein-bound dopamine. Biochemistry. 2006;45:15120–15128. doi: 10.1021/bi0614434. [DOI] [PubMed] [Google Scholar]
- 28.Traut RR, Bollen A, Sun TT, Hershey JW, Sundberg J, Pierce LR. Methyl 4-mercaptobutyrimidate as a cleavable cross-linking reagent and its application to the Escherichia coli 30S ribosome. Biochemistry. 1973;12:3266–3273. doi: 10.1021/bi00741a019. [DOI] [PubMed] [Google Scholar]
- 29.Palacian E, Gonzalez PJ, Pineiro M, Hernandez F. Dicarboxylic acid anhydrides as dissociating agents of protein-containing structures. Mol Cell Biochem. 1990;97:101–111. doi: 10.1007/BF00221051. [DOI] [PubMed] [Google Scholar]
- 30.Gibbons I, Perham RN. The reaction of aldolase with 2-methylmaleic anhydride. Biochem J. 1970;116:843–849. doi: 10.1042/bj1160843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li SW, Lin TS, Minteer S, Burke WJ. 3,4-Dihydroxyphenylacetaldehyde and hydrogen peroxide generate a hydroxyl radical: possible role in Parkinson's disease pathogenesis. Brain Res Mol Brain Res. 2001;93:1–7. doi: 10.1016/s0169-328x(01)00120-6. [DOI] [PubMed] [Google Scholar]
- 32.Lamensdorf I, Eisenhofer G, Harvey-White J, Nechustan A, Kirk K, Kopin IJ. 3,4-Dihydroxyphenylacetaldehyde potentiates the toxic effects of metabolic stress in PC12 cells. Brain Res. 2000;868:191–201. doi: 10.1016/s0006-8993(00)02309-x. [DOI] [PubMed] [Google Scholar]
- 33.Bindoli A, Rigobello MP, Deeble DJ. Biochemical and toxicological properties of the oxidation products of catecholamines. Free Radic Biol Med. 1992;13:391–405. doi: 10.1016/0891-5849(92)90182-g. [DOI] [PubMed] [Google Scholar]
- 34.Mattammal MB, Strong R, Lakshmi VM, Chung HD, Stephenson AH. Prostaglandin H synthetase-mediated metabolism of dopamine: implication for Parkinson's disease. J Neurochem. 1995;64:1645–1654. doi: 10.1046/j.1471-4159.1995.64041645.x. [DOI] [PubMed] [Google Scholar]
- 35.Eisenhofer G, Aneman A, Hooper D, Holmes C, Goldstein DS, Friberg P. Production and metabolism of dopamine and norepinephrine in mesenteric organs and liver of swine. Am J Physiol. 1995;268:G641–649. doi: 10.1152/ajpgi.1995.268.4.G641. [DOI] [PubMed] [Google Scholar]
- 36.Burke WJ, Kumar VB, Pandey N, Panneton WM, Gan Q, Franko MW, O'Dell M, Li SW, Pan Y, Chung HD, Galvin JE. Aggregation of alpha-synuclein by DOPAL, the monoamine oxidase metabolite of dopamine. Acta Neuropathol. 2008;115:193–203. doi: 10.1007/s00401-007-0303-9. [DOI] [PubMed] [Google Scholar]
- 37.Mazzola JL, Sirover MA. Alteration of intracellular structure and function of glyceraldehyde-3-phosphate dehydrogenase: a common phenotype of neurodegenerative disorders? Neurotoxicology. 2002;23:603–609. doi: 10.1016/s0161-813x(02)00062-1. [DOI] [PubMed] [Google Scholar]
- 38.Kagan VE, Tyurina YY. Recycling and redox cycling of phenolic antioxidants. Ann N Y Acad Sci. 1998;854:425–434. doi: 10.1111/j.1749-6632.1998.tb09921.x. [DOI] [PubMed] [Google Scholar]
- 39.Lane CF. Sodium cyanoborohydride - highly selective reducing agent for organic functional groups. Synthesis-Stuttgart. 1975:135–146. [Google Scholar]