Figure 2.
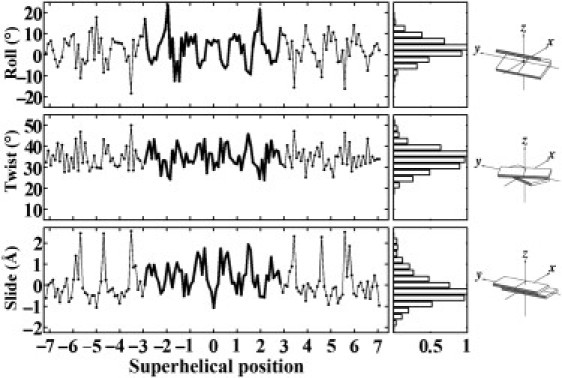
Variation of the three primary basepair step parameters (Roll, Twist, Slide), computed with 3DNA (46,47), as a function of superhelical position of DNA on the 147-bp nucleosome core-particle structure (NDB_ID pd0287) (2). The step parameters of the 60 dimer steps (61 bp) bound to the (H3·H4)2 tetramer are highlighted by heavy lines at the center of the plotted data. Superhelical positions correspond to the number of double-helical turns a dimeric step is displaced from the structural dyad on the central basepair (denoted by 0). Histograms on the right edge of the figure are derived from the distribution of step parameters in 135 other well-resolved protein-DNA complexes (see Methods and Supporting Material) and scaled with respect to a value of unity for the most populated parametric ranges. The fixed angular scales emphasize the preferential deformation of nucleosomal DNA via bending rather than twisting, compared to the similar ranges of Roll and Twist in other protein-DNA complexes. Block images of step parameters obtained with 3DNA (46,47) and illustrated with RasMol (87).
