Figure 4.
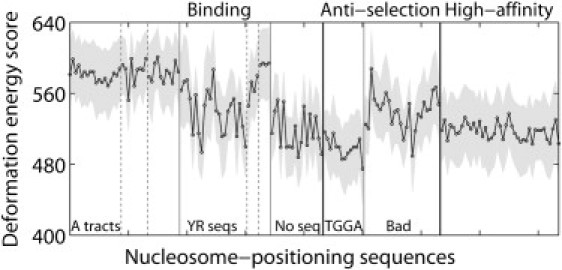
Cost of threading 88 known binding sequences from the mouse genome (9), 40 sequences refractory to nucleosome formation (anti-selection) (24), and 41 high-affinity nucleosome-binding fragments (22) on the central 60 basepair steps of the DNA in the 147-bp nucleosome core-particle structure (NDB_ID pd0287) (2). Sequences are arranged in the order reported in Table S2 and grouped according to literature descriptors (9,22,24). Thin vertical lines highlight sequences of similar types. Total threading scores—mean values 〈U〉 (points connected by dashed lines) and standard deviations σU (values equal to half the width of the gray corridors containing the points) for all possible settings of each sequence on the structural template—based on six-parameter elastic functions for dimer steps from all conformational categories, i.e., the A + B + AB data set in Table 1. See Table S5 for the scores of individual fragments.
