Abstract
The development of genetic detection protocols for field applications is an important aspect of modern medical diagnostic technology and environmental monitoring. In this paper, we report a rapid, portable, and inexpensive DNA hybridization technique using a bead-based microfluidic platform that functions by passing fluorescently labeled target DNA through a chamber packed with functionalized beads within a microfluidic channel. DNA hybridization is then assessed using a digital camera attached to a Clare Chemical DR-45M dark reader non-UV transilluminator that uses visible light as an excitation source and a blue and amber filter to reveal fluorescence. This microfluidic approach significantly enhances hybridization by reducing the diffusion time between target DNA and the silica surface. The use of probe-functionalized beads as solid support also enhances the sensitivity and limit of detection due to a larger surface area per unit volume. This platform could be adapted for use in medical applications and environmental monitoring, including the detection of harmful organisms in the ballast water of ships.
INTRODUCTION
The development of rapid detection techniques for bacteria, viruses, other harmful species, or species of interest in environmental field surveys is of immense importance for the advancement of modern medical diagnostic technology and environmental monitoring. Market demands for a portable device for field applications would likely be high because these are necessary for world health, epidemic control, environmental monitoring, and antiterrorism∕biowarfare applications. Several excellent reviews of pathogen identification through diverse nucleic acid based assays, including microarray and real time polymerase chain reaction (PCR), can be found in literature.1, 2 Conventional microarray analyses, however, involve a time-consuming and expensive detection protocol that often results in large errors and inconsistent results.3 The process requires a series of mixing, incubation, and washing steps. A single genetic assay of this type can take from 15 to 24 h. This is because of the rate-limiting diffusion step and is attributed to insufficient mass transport of the target DNA from the solution to reach the localized oligomer sequences printed at specific pixels on the surface of a microarray slide.4, 5 Both microarray and real time PCR are costly and are not very portable. Therefore, they are not convenient for field applications. Thus, there is a growing need for a rapid, portable device for field-based species detections.
The development of portable devices in conjunction with microfluidic and lab-on-a-chip platforms for biological applications has shown significant advancement during the past decades.6, 7 Portable microfluidic devices could, thus, have considerable impact on rapid diagnostics, especially for field use or at any point of need.8 Integration of such a platform on a microchip would enhance the reaction efficiency, simplify detection protocol, reduce detection time, and lower the quantity of loading samples. Herein, we report a rapid, robust, and simple genetic detection assay system on a bead-based microfluidic platform where the probe-functionalized beads are used as a solid support to detect fluorescently tagged target DNA. The fluorescent intensity is captured by using a digital camera attached to a Clare Chemical DR-45M dark reader non-UV transilluminator that employs visible light as the excitation source accompanied by a blue and amber filter.
A number of research groups have been actively working with bead-based microanalytical systems. Fan et al.9 reported DNA hybridization in a microfluidic array using paramagnetic beads as a transportable solid support. A capillary platform for DNA analysis was demonstrated by lining up individual beads within a capillary tube.10 DNA hybridization using chip-based sensor arrays composed of individual addressable microbeads has also been reported.11 Recently, the use of microbead arrays was utilized in gene expression analyses.12 Other interesting studies of bead-based microfluidic biosensors are well documented in literature.13, 14
The use of beads as solid support to capture the DNA in a microfluidic chamber has a number of advantages. First, the bead’s surface to volume ratio is significantly higher than any flat surface or the interior surface of the microfluidic channel and this provides more hybridization sites for target DNA, thus enhancing sensitivity and the limit of detection for assays based on immobilized probes. Additionally, the use of beads offers a simple way to modify and characterize surfaces. Finally, the beads also enhance the effective mixing of solutions in the microfluidic system.15
In the present study, we describe the development of a continuous flow system on a bead-based microfluidic platform for rapid genetic identification. We describe an initial application to detect the presence of green crab (Carcinus maenas), a marine crab that has often been economically and environmentally damaging where it has been introduced, often via the ballast water of ships.16, 17, 18 In this approach, the probe-functionalized beads (∼106 beads) are trapped within an 8×108 μm3 small chamber in the microfluidic channel and the fluorescently labeled target DNA is exposed to the packed bead chamber for rapid hybridization. This reduces the diffusion rate between DNA and probes functionalized on the bead surface as DNA has to travel smaller distances to encounter probes due to the close packing of beads within a small chamber. Furthermore, the trapping of beads in the microfluidic channel also enhances detection sensitivity by concentrating the fluorescence intensity within a small chamber. Thus, the integration of such a microfluidic platform with a bead-based approach allows rapid hybridization and washing, decreasing the overall detection time to 1 h. Green crab DNA was used as the target species to study the genetic detection on this bead-based microfluidic platform, with a green crab congener, Carcinus aestuarii, used as a control to test the species specificity of the approach.
EXPERIMENTAL DETAILS
The bead-based microfluidic detection platform consists of five steps: (i) microchip fabrication on glass slides, (ii) fabrication of filters within the microchannel using a mixture of methyacrylate photopolymers, (iii) functionalization of the oligomer probe on beads, (iv) asymmetric PCR, and (v) a microchip bead-based hybridization assay. Each step is described briefly in Sec. 2A.
Microchannel fabrication
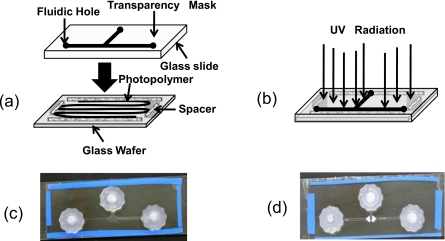
A microfluidic cassette with a T-shaped microfluidic channel network [shown in Fig. 1c] is constructed using a liquid phase photopolymerization technique previously described in literature.19 The top and bottom surfaces of the cassettes are made up of glass slides, and the side walls of the channels are made out of acrylate polymer. The process consists of the following steps.
-
(a)
Step 1 [Fig. 1a]. A microscopic glass slide is used to construct the upper portion of the chip. The holes are drilled at defined locations for fluidic inlet and outlet ports and a mask (printed on a transparency sheet) is attached to one of the surfaces.
-
(b)
Step 2 [Fig. 1a]. The lower portion of the chip is formed with a glass wafer and the four edges of one surface are taped, and a layer of monomer (Impruv® 363, Loctite Inc.; Rocky Hill, CT) is spread over it. The tape acts a spacer between the two slides and represents the height of the channel. The thickness of the tape used is around 200 μm.
-
(c)
Step 3 [Fig. 1b]. The upper layer is then put over the monomer glass wafer and is subjected to free-radical polymerization by irradiating the system for 5 s in a Electro-cure 500 UV curing chamber 81432 (Electro-lite Corp., Bethel, CT) with four 15 W fluorescent blacklight tubes, producing UV light of predominantly 365 nm wavelength at an intensity of 30 mW∕cm2 in the polymerization zone. Clear areas of the mask allow transmission of the UV light, curing the monomer while dark areas of the mask block the UV light and leave regions of unpolymerized monomer.
-
(d)
Step 4 [Fig. 1c]. The mask is removed, and the unpolymerized monomer is washed out with acetone to develop the desired channel. Finally, the chip is exposed in an UV curing chamber for another 2 min and the fluidic connectors are attached. The fabricated T-shaped microchannel has dimensions of 7×0.02×30 cm3 in width(cm)×height(cm)×length(cm), respectively.
Figure 1.
Fabrication of the microchannel prototype. (a) The upper layer consists of a transparency mask, a glass slide with fluidic holes drilled in. The lower layer consists of monomer solution poured on top of a tapped glass wafer. (b) The two layers are combined and exposed to UV light from above, curing only the areas not protected by the mask. (c) The picture of the chip after fabrication of the microchannel. (d) The picture of the chip after formation of the polymeric filter within the microfluidic channel.
Fabrication of filters within microchannel
Filters with pore dimension of ∼2–3 μm are fabricated on either side of the horizontal arm of the T-junction to create a region where 10 μm DNA-coated beads, when loaded through the vertical arm, can be trapped [Fig. 1d]. The on-chip fabrication of the filter was achieved by modifying a previous technology used to fabricate porous monoliths.20
In brief, a mixture of trimethylolpropane trimethacrylate and 2,3-epoxypropyl methacrylate (glycidyl methacrylate) in the ratio of 7:3 is used as monomer and another mixture of toluene and 2,2,4-trimethylpentane in the ratio of 4:6 is used as porogen. 0.35 mg benzoin methyl ether, a photoinitiator, is then added to a mixture consisting of 0.4 ml of monomer and 0.6 ml of porogen. The resulting solution is then loaded into the microchannels. Using the same setup that was used for fabricating the microchannel network, but with a new mask that allows light to pass through only those regions in which the filter is desired, free-radical photopolymerization is initiated and allowed to proceed for 68 s. Due to the presence of porogens (inert organic compounds), a spongy mass, as opposed to a solid polymer, is obtained from the reaction. When methanol is fed through the vertical arm at a low flow rate (3 ml∕h), the porogen, unreacted monomers, and some partially reacted monomers within the porous mass are washed away, but the more highly polymerized material remains, yielding a porous monolith within the microfluidic channel. The size of the pores within the monolith can be tuned by changing the ratio of the compounds constituting the porogen, the volume fraction of the porogens, and the amount of photoinitiator used.21
Functionalization of probe on bead surface
To hybridize target DNA on the bead-based microfluidic platform, a species-specific amino functionalized oligonucleotide primer (27-mer), complementary to the target DNA, is attached to 10 μm carboxylated silica beads. This is achieved by coupling –COOH and –NH2 groups with mixture of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide to produce DNA attached silica bead by amide linkages and is performed as reported in literature.22
Asymmetric PCR amplification
To simplify the bead-based hybridization platform and also to avoid the denaturation of the target DNA during the hybridization step in the experimental setup (i.e., to prevent the denatured dsDNA from recombining before it reaches and interacts with probe-functionalized silica beads within the microchannel), asymmetric PCR is performed to produce ssDNA. In this approach, an unequal concentration of primers is used; otherwise it is similar to standard, symmetric PCR that produces dsPCR product. Initially, amplification starts exponentially, but as the lower concentrated primer is exhausted, the higher concentrated primer continues to amplify only one strand of DNA, thus producing ssDNA. An asymmetric PCR reaction amplifying cytochrome c oxidase subunit I (COI) was performed using a fluorescently labeled forward PCR primer (0.4 mM concentration) and a reverse, nonfluorescent primer (0.8 nM concentration) to amplify ∼500 base pair green crab ssDNA to study on-chip DNA hybridization. Similarly, the congeneric species Carcinus aestuarii was amplified and tested as a negative control.
Procedure for microchip-bead-based hybridization assay
The first step of the hybridization experiment is to pass 2% serum bovine albumin solution through the microchannel to prevent any nonspecific binding of target DNA or fluorescent dye to the filters and the surfaces of the channels. The probe-functionalized silica beads are then passed through the vertical fluidic inlet of the microchannel to trap the beads within the small chamber and then a 4X SSC hybridization buffer is passed through the channel to make sure that the beads are closely packed within the chamber. After successful packing of the beads, 100 μl of a solution of the fluorescently labeled ssDNA is flown through at 50 °C at a very slow flow rate of 0.5 ml∕h for hybridization. De-ionized water is then passed through the channel to remove all nonspecific DNA bound to the surface of the beads and∕or to the filters. Finally, the fluorescence of the packed bead chamber is quantified using a digital camera attached to a Clare Chemical DR-45M dark reader non-UV transilluminator that uses visible light source for fluorescence detection. The dark reader uses two filters (blue and amber filter) to reveal fluorescence. The blue filter placed in between the light source and the sample allows only blue excitation light to pass through it. The amber filter is then placed between the sample and the observer, which removes the blue incident light but allows passage of the red and green fluorescent components. This emits the wavelength in the range of 400–500 nm and falls in the excitation region of fluorophor Alexa488, which is used to detect DNA hybridization. A confocal laser scanning microscope (Zeiss LSM Pascal 5) is also used to further verify on-chip hybridization.
RESULTS AND DISCUSSIONS
This approach is rapid: it took about 1 h from introducing the DNA into the channel to the completion of optical quantification of the results. Detection sensitivity is in the range of 100 pM.
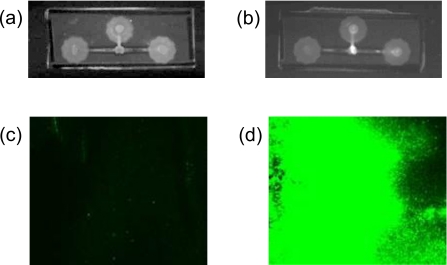
Figure 2a shows the image of the control chip with bare silica beads and Fig. 2b shows the image of the experimental chip with probe-functionalized silica beads packed within the microfluidic channel after passing the fluorescently labeled target green crab ssDNA through the bead chamber, respectively. A bright white spot can be seen in the packed bead chamber of the experimental chip, whereas no such brightness is observed in the control chip. This observation clearly suggests that the fluorescently labeled ssDNA is hybridized with probes attached to the surface of the silica beads. Further evidence for on-chip hybridization is provided by laser scanning confocal microscope. Figures 2c, 2d show the fluorescence images recorded by laser scanning of the packed silica beads of the control and experimental chip, respectively. The presence of intense fluorescence in the experimental chip further confirms the successful hybridization of the target green crab ssDNA on probe-functionalized silica beads trapped within a small microfluidic chamber. No such appreciable fluorescence is observed for the control chip. The rapid detection of DNA hybridization on this microfluidic platform clearly suggests the packing of beads within a small chamber and the passing of target DNA through it significantly reduces the diffusion time between target DNA and bead surface compare to microarray-based detection technique. Moreover, the large surface area to volume ratio of such beads allows a microreservoir to capture a high concentration of the target DNA sample with picomolar sensitivity. As all fluorescently labeled DNA are concentrated within a microreservoir after DNA hybridization, the fluorescent intensity of the chamber is very high. As a reference, a single pixel on a DNA microarray has an area of 10−2 cm2. In contrast, the area of a chamber filled with a microliter of microbeads (50% by volume) is four orders of magnitude higher, with a proportionally larger fluorescent intensity.
Figure 2.
The image of (a) the control chip with bare silica beads and (b) the experimental chip with probe-functionalized silica beads packed within a small chamber in the microfluidic channel after passing fluorescently labeled target green crab ssDNA through both the chips, respectively, is recorded using a digital camera attached to a Clare Chemical DR-45M dark reader. (c) The fluorescence image of the control and (d) the experimental chip recorded by a confocal laser scanning microscope.
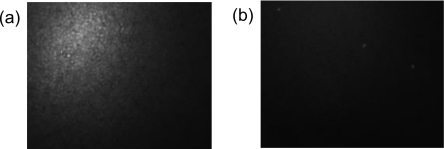
To demonstrate the specificity of this bead-based microfluidic platform for genetic identification, the following experiments are conducted. A comparative test between the green crab and the closely related congeneric species Carcinus aestuarii is performed. The experiments involved passing amplified green crab and its congener ssDNA through two separate chips packed with probe-functionalized (green crab specific) beads. After successful packing of beads, the fluorescently labeled COI PCR product from both the green crab (C. maenas) and its congener (C. aestuarii) is run through two separate chips at 50 °C with a very slow flow rate of 0.5 ml∕h. The chips were then washed with de-ionized water to remove the nontarget DNA product that is bound either to the surface of the beads or to the filter. The fluorescence of the packed bead chamber was then assessed using a fluorescent microscope (Fig. 3). High fluorescence intensity was observed in the green crab chip [Fig. 3a], and no appreciable fluorescence was observed for the congeneric species chip [Fig. 3b], despite the fact that the probe used to detect the green crab PCR product differs by only three mismatched bases from the congeneric DNA sequence. This observation clearly demonstrates the high specificity of bead-based detection chip toward a particular species and also demonstrates that the bead-based microfluidic platform can successfully detect a minimum of three base mismatches.
Figure 3.
(a) The image of the probe-functionalized beads trapped within a microfluidic channel after passing fluorescently labeled (a) green crab ssDNA and (b) its congener ssDNA, respectively, is captured using a fluorescent microscope.
In conclusion, we have successfully demonstrated a rapid, simple, and portable genetic identification detection technique on a bead-based microfluidic platform. The fabrication of this platform is very simple and the use of a digital camera accompanied with a visible light source with appropriate filters for DNA hybridization detection makes the platform simple, portable, and cost effective. The detection sensitivity can be further improved by using a more powerful and expensive optical system, although it may limit its portability, an advantage of the reported microfluidic detection platform. Further, the detection platform is based on a continuous flow system that allows the passage of a large sample volume of low DNA concentration for longer time, thus improving sensitivity at the expense of detection time. With the proper integration of a modified PCR chip, hybridization steps, and the appropriate microfluidic fabrication techniques, this platform then could be introduced as a first single-target prototype for a rapid field-use genetic diagnostic kit. Quantifying the target DNA (as opposed to measuring its presence or absence) is not currently possible but a rapid and portable positive-negative diagnostic platform would find many important field-use applications as a preliminary screening step, such as epidemic control at ports∕airports, avian flu monitoring of poultry imports, and environmental monitoring.
ACKNOWLEDGMENTS
We would like to thank Matthew Barnes and Sagnik Basuray for their valuable contributions. This work was financially supported by the Great Lake Protection Fund.
References
- Barken K. B., Haagensen J. A. J., and Tolker-Nielsen T., Clin. Chim. Acta 384, 1 (2007). 10.1016/j.cca.2007.07.004 [DOI] [PubMed] [Google Scholar]
- Mothershed E. A. and Whitney A. M., Clin. Chim. Acta 363, 206 (2006). 10.1016/j.cccn.2005.05.050 [DOI] [PubMed] [Google Scholar]
- Tu I. P., Schaner M., Diehn M., Sikic B. I., Brown P. O., Botstein D., and Fero M. J., BMC Genomics 5, 64 (2004). 10.1186/1471-2164-5-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kian-Kok Ng J., Feng H., and Liu W. T., Anal. Chim. Acta 582, 295 (2007). 10.1016/j.aca.2006.09.016 [DOI] [PubMed] [Google Scholar]
- Liu R. H., Lenigk R., Druyor-Sanchez R. L., Yang J., and Grodzinski P., Anal. Chem. 75, 1911 (2003). 10.1021/ac026267t [DOI] [PubMed] [Google Scholar]
- Verpoorte E., Lab Chip 3, 60N (2003). 10.1039/b313217j [DOI] [PubMed] [Google Scholar]
- Vilkner T., Janasek D., and Manz A., Anal. Chem. 76, 3373 (2004). 10.1021/ac040063q [DOI] [PubMed] [Google Scholar]
- Tudos A. J., Besselink G. A., and Schasfoort R. B. M., Lab Chip 1, 83 (2001). 10.1039/b106958f [DOI] [PubMed] [Google Scholar]
- Fan Z. H., Mangru S., Granzow R., Hcancy P., Ho W., Dong Q., and Kumar R., Anal. Chem. 71, 4851 (1999). 10.1021/ac9902190 [DOI] [PubMed] [Google Scholar]
- Kohara Y., Anal. Chem. 75, 3079 (2003). 10.1021/ac0341214 [DOI] [PubMed] [Google Scholar]
- Ali M. F., Kirby R., Goodey A. P., Rodriguez M. D., Ellington A. D., Neikirk D. P., and McDevitt J. T., Anal. Chem. 75, 4732 (2003). 10.1021/ac034106z [DOI] [PubMed] [Google Scholar]
- Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D. H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Roth R., George D., Eletr S., Albrecht G., Vermaas E., Williams S. R., Moon K., Burcham T., Pallas M., DuBridge R. B., Kirchner J., Fearon K., Mao J., and Corcoran K., Nat. Biotechnol. 18, 630 (2000). 10.1038/76469 [DOI] [PubMed] [Google Scholar]
- Smistrup K., Kjeldsen B. G., Reimers J. L., Dufva M., Petersen J., and Hansen M. F., Lab Chip 5, 1315 (2005). 10.1039/b510995g [DOI] [PubMed] [Google Scholar]
- Goral V. N., Zaytseva N. V., and Baeumner A. J., Lab Chip 6, 414 (2006). 10.1039/b513239h [DOI] [PubMed] [Google Scholar]
- Lund-Olesen T., Dufva M., and Hansen M. F., J. Magn. Magn. Mater. 311, 396 (2007). 10.1016/j.jmmm.2006.10.1171 [DOI] [Google Scholar]
- Deagle B. E., Bax N., Hewitt C. L., and Patil J. G., Mar. Freshwater Res. 54, 709 (2003). 10.1071/MF03031 [DOI] [Google Scholar]
- Patil J. G., Gunasekera R. M., Deagle B. E., Bax N. J., and Blackburn S. I., Biol. Invasions 7, 983 (2005). 10.1007/s10530-004-3119-8 [DOI] [Google Scholar]
- Patil J. G., Gunasekera R. M., Deagle B. E., and Bax N. J., Mar. Biotechnol. 7, 11 (2005). 10.1007/s10126-004-0034-z [DOI] [PubMed] [Google Scholar]
- Sengupta S., Ziaie B., and Barocas V. H., Sens. Actuators B 99, 25 (2004). 10.1016/j.snb.2003.08.017 [DOI] [Google Scholar]
- Viklund C., Ponten E., Glad B., and Irgum K., Chem. Mater. 9, 463 (1997). 10.1021/cm9603011 [DOI] [Google Scholar]
- Senagupta S. and Chang H. -C., Encyclopedia of Microfluidics and Nanofluidics (Springer-Verlag, New York, 2008), p. 1128. [Google Scholar]
- Marquette C. A. and Blum L. J., Top. Curr. Chem. 261, 113 (2006). [Google Scholar]