Abstract
Antigenic variation allows Trypanosoma brucei to evade the host immune response by switching the expression of 1 out of ∼15 telomeric variant surface glycoprotein (VSG) expression sites (ESs). VSG ES transcription is mediated by RNA polymerase I in a discrete nuclear site named the ES body (ESB). However, nothing is known about how the monoallelic VSG ES transcriptional state is maintained over generations. In this study, we show that during S and G2 phases and early mitosis, the active VSG ES locus remains associated with the single ESB and exhibits a delay in the separation of sister chromatids relative to control loci. This delay is dependent on the cohesin complex, as partial knockdown of cohesin subunits resulted in premature separation of sister chromatids of the active VSG ES. Cohesin depletion also prompted transcriptional switching from the active to previously inactive VSG ESs. Thus, in addition to maintaining sister chromatid cohesion during mitosis, the cohesin complex plays an essential role in the correct epigenetic inheritance of the active transcriptional VSG ES state.
Introduction
The parasite Trypanosoma brucei undergoes different developmental stages between mammalian host and tsetse vector during its life cycle. In the mammalian host, extracellular bloodstream form parasites elude the host immune response by periodically changing their main surface antigen, the variant surface glycoprotein (VSG). To achieve the expression of a single type of VSG on the surface, only 1 out of ∼15 possible subtelomeric VSG expression sites (ESs) is expressed at a given time (Borst and Ulbert, 2001). Unusually, the active VSG ES is transcribed by RNA polymerase I (Gunzl et al., 2003) and is located in an extranucleolar position (Chaves et al., 1998) associated with an extranucleolar body named the ES body (ESB; Navarro and Gull, 2001).
Transcriptional switching of the active VSG ES occurs at a very low frequency and does not involve DNA sequence changes at the promoter region; thus, transcriptional regulation of VSG ESs is probably epigenetic. It has been suggested that maintenance of the epigenetic state of monoallelic expression of the active VSG ES could be mediated by exclusive association to the ESB (Navarro and Gull, 2001; Borst, 2002; Navarro et al., 2007). If this were the case, the propagation of a stable, active VSG ES–ESB complex must be coordinated with chromosome duplication and segregation. Therein, to understand how the transcriptional state of VSG ESs can be inherited in the next generation, it is important to analyze how the active VSG ES–ESB complex behaves throughout the cell cycle. This is particularly important from S to M phases, when new sister chromatids of the active and inactive VSG ESs are synthesized, likely epigenetically labeled, and segregated to give rise to progeny with the same VSG ES transcriptional configuration.
In eukaryotes, sister chromatids remain together after DNA replication until mitosis in a process that has been shown to be essential for the correct inheritance of genetic material. A large, ringlike multisubunit protein complex called cohesin serves to link the two sister chromatids together. The cohesin complex consists of four major subunits, SMC1, SMC3, SCC3, and SCC1, the last of which is cleaved by separase to release duplicated sister chromatids during the metaphase–anaphase transition (Uhlmann, 2001b; Nasmyth, 2002). Although new evidence is emerging suggesting that the cohesin complex is involved in the regulation of gene expression in diverse organisms such as yeast, invertebrates, and mammals, the molecular mechanisms underlying this regulation remain largely unknown (for reviews see Losada, 2007; Peric-Hupkes and van Steensel, 2008). In trypanosomes, recent data support a conserved function of the cohesin complex in the segregation of chromosomes (Gluenz et al., 2008; Sharma et al., 2008; Bessat and Ersfeld, 2009). However, a noncanonical role for the cohesion complex in the regulation of gene expression remains completely unexplored in trypanosomes.
In this study, we investigate how the association of the active VSG ES to the extranucleolar pol I–containing ESB is passed from one generation to the next. We show that sister chromatids of the active VSG ES remain held together longer than other loci and that they stay in association with the unique ESB until chromosome segregation. Compromising the integrity of the cohesin complex results in the loss of association between the active VSG ES and the ESB as well as triggers a transcriptional switch from the active VSG ES to previously inactive ones.
Results
The ESB remains a single nuclear structure throughout S and G2 phases of the cell cycle
In T. brucei, duplication of the mitochondrial genome (kinetoplast [K]) antecedes nuclear (N) mitosis, allowing for facile tracking of individual cells at different stages of the cell cycle (Woodward and Gull, 1990; Siegel et al., 2008). Thus, trypanosomes with one kinetoplast and one nucleus (1K1N) are known to correspond to nuclear interphase cells (G1-synthesis). Later, trypanosomes displaying two kinetoplasts and one nucleus (2K1N) correspond to nuclear G2 and mitotic phases. Upon nuclear mitosis, cells show a 2K2N configuration, which persists until cytokinesis is finished. The active VSG ES is associated with the ESB in both 1K1N (G1-S) and 2K2N (postmitotic) cells (Navarro and Gull, 2001). Thus, it has been hypothesized that the exclusive association of the active VSG ES to the single ESB underlies the mechanism by which monoallelic expression of VSG ESs is achieved (Navarro and Gull, 2001; Borst, 2002). However, nothing is currently known about how the association of the active VSG ES with the ESB is passed on to the next generation.
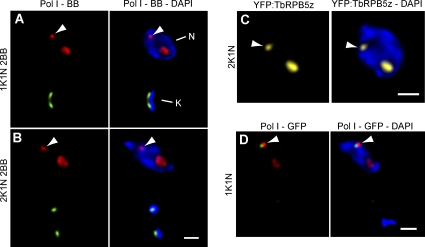
To gain insight into this question, we first determined whether ESB duplication occurs upon synthesis of a newly replicated active VSG ES. As a marker for nuclear S phase, we used basal bodies (BBs; Woodward and Gull, 1990). 3D immunofluorescence (IF; Landeira and Navarro, 2007) analysis using anti-TbRPA1, the largest subunit of RNA polymerase I (Navarro and Gull 2001), recognized the bulk of pol I resident in the nuclear region exhibiting faint DAPI-stained DNA corresponding to the nucleolus as well as the pol I–containing extranucleolar ESB (Fig. 1 A). It has been previously described that the ESB is detected in ∼60% of 1K1N cells (Navarro and Gull 2001). Similarly, in 50 ± 6% of S-phase cells (1K1N and 2BB), a single ESB was clearly detected in three different cells lines analyzed by 3D IF (Fig. 1 A). Likewise, in G2-mitotic cells showing a 2K1N configuration, the ESB was detected in 57 ± 8% of cells (Fig. 1 B). Importantly, we were not able to detect a significant percentage of cells displaying two ESBs in either S (0.9 ± 1%; P < 0.001) or G2 phase (0.6 ± 0.8%; P < 0.001).
Figure 1.
The ESB remains a single nuclear structure throughout S and G2 phases of the cell cycle. (A and B) Bloodstream form trypanosomes were subjected to double indirect 3D IF using polyclonal anti-TbRPA1 antibodies, monoclonal antibodies against the BB (BBA4), and DAPI staining. Cells of three different cell lines were grouped as defined by the number of nuclei (N), kinetoplasts (K), and BBs. 1K1N 2BB (n = 50) and 2K1N 2BB (n = 50) trypanosomes were scored for the number of extranucleolar pol I–containing ESBs. Representative 1K1N 2BB cells (A) and 2K1N 2BB (B) cells are shown as maximum intensity projections of three-channel 3D stacks showing anti–pol I signal in red, BBA4 signal in green, and DAPI staining in blue. (C) Maximum intensity projections of a two-channel 3D stack showing direct fluorescence of the fusion protein YFP-TbRPB5z in yellow and DAPI staining in blue. (D) Double indirect 3D IF of a cell line GFP tagged in the active VSG ES. Maximum intensity projections of a two-channel 3D stack showing polyclonal anti-TbRPA1 antibodies (red), monoclonal antibodies against GFP (green), and DAPI staining (blue) are shown. (A–D) Arrowheads indicate the ESB. Bars, 1 µm.
To analyze ESB dynamics during G2 phase using a different approach, we obtained transformed trypanosomes expressing an N-terminal YFP-TbRPB5z fusion, which was previously shown to render a functional protein that is a component of both the nucleolus and the ESB in bloodstream trypanosomes (Devaux et al., 2007). Consistent with the 3D IF analysis in Fig. 1 B, using the YFP-TbRPB5z fusion protein, DAPI staining, and 3D analysis, we were not able to detect two ESBs within an individual nucleus in 2K1N cells; however, a single ESB was clearly detectable in a high percentage of cells (55%; Fig. 1 C).
Investigation of ESB dynamics is facilitated by its extranucleolar position; however, the lack of a specific marker exclusively present in the ESB has hindered some experiments. We could efficiently discern the ESB from other pol I–containing areas in the nucleus by using GFP chromosome tagging of the active VSG ES promoter sequences, which exclusively associate with the ESB (Navarro and Gull, 2001). We investigated whether ESB duplication occurs in S phase. Among cells in which we successfully identified the signal coming from the GFP-tagged active VSG ES associated to extranucleolar pol I signal, 97% of cells showed a single, extranucleolar pol I signal, which is consistent with the existence of a single ESB upon DNA replication (Fig. 1 D). Altogether, these data suggest that the ESB remains a single nuclear structure during S and G2 phases.
The active VSG ES displays delayed separation of sister chromatids
We next analyzed the dynamics of sister chromatids of the active VSG ES during cell cycle progression. GFP tagging of chromosomes has been previously used to detect the separation of sister chromatids, as indicated by a two GFP–dot signal (Belmont, 2001; McKee, 2004). We first established the general dynamic of sister chromatids of a pol I–transcribed locus at different stages of the cell cycle in the bloodstream form of T. brucei. We analyzed sister chromatid separation by IF using anti-GFP antibodies for one of several ribosomal DNA (rDNA) loci tagged with GFP-LacI, as described previously (Landeira and Navarro, 2007). We grouped the population into 1K1N (G1 and S), 2K1N (G2 and mitotic), and 2K2N (postmitotic) cells and scored whether one or two GFP dots were detectable. The vast majority of interphase cells (1K1N) displayed a single GFP dot (Fig. S1, A and C), whereas only 5% of cells showed two GFP dots (Fig. S1 A). Upon cell cycle progression, a high proportion of cells showed a single GFP dot in G2 and mitotic phases (2K1N; 64%; Fig. S1, A and D), suggesting that after DNA synthesis, sister chromatids are held together in T. brucei, similar to what occurs in other eukaryotes. However, we easily detected separation of sister chromatids in 36% of 2K1N nuclei, suggesting that sister chromatids normally separate during premitotic and mitotic phases in T. brucei (Fig. S1, A, E, and F). Postmitotic 2K2N cells showed two postmitotic nuclei displaying one GFP dot each (Fig. S1, A and G). Similar results were obtained when we analyzed the separation of sister chromatids using cell lines GFP tagged in different loci in both bloodstream and procyclic developmental forms (Fig. S1 A).
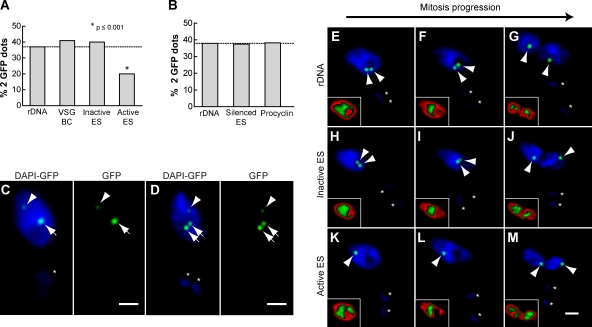
In contrast, analysis of sister chromatid dynamics during the cell cycle for the active VSG ES locus suggested a distinct pattern in 2K1N cells because two GFP dots were detected in a lower percentage in this group of cells (Fig. S1 B). Thus, we decided to expand the number of 2K1N (G2 and mitotic phases) cells analyzed. Interestingly, the frequency of detection of separated sister chromatids of the active VSG ES promoter was only 20%, which is significantly lower than the 37–41% detected for control loci, which included an rDNA locus, an inactive VSG ES promoter, and the basic copy (BC) of a VSG (VSG121-BC; P < 0.001; Fig. 2 A). In procyclic form trypanosomes, where no VSG is expressed, sister chromatids of the VSG ES did not exhibit a difference in their separation when compared with other pol I–transcribed loci (∼37%; Fig. 2 B). Furthermore, the procyclin locus did not show a difference in the separation of sister chromatids when compared with the rDNA locus (38–37%) in the procyclic form (Fig. 2 B). The high similarity in the proportion of cells showing separation of sister chromatids among the seven studied loci (37–41%), with the exception of the active VSG ES (20%; Fig. 2, A and B), suggests that the sister chromatids of the active VSG ES locus may be targeted for enhanced cohesion.
Figure 2.
Enhanced sister chromatid cohesion is a distinctive feature of the active VSG ES. Different cell lines with GFP-tagged loci described previously (Landeira and Navarro, 2007) were subjected to indirect IF using polyclonal anti-GFP antibodies and DAPI staining. (A) Histogram showing the percentage of nuclei containing two GFP dots in 2K1N bloodstream form trypanosomes with a GFP tag in either the rDNA locus (n = 196), VSG-BC (n = 166), inactive VSG ES (n = 104), or active VSG ES (n = 161). (B) Histogram showing the percentages of cells containing two GFP dots in 2K1N procyclic form trypanosomes with a GFP tag in either the rDNA locus (n = 166), silenced VSG ES (n = 184), or procyclin locus (n = 170). (A and B) Dashed lines indicate the value of the rDNA that was used as the expected value in Fisher's exact test. (C and D) Representative pictures of a 1K1N (C) or 2K1N (D) cell showing GFP dot detection in a double GFP-tagged bloodstream cell line, with GFP marking the rDNA locus (250 lac operator repeats; arrows) and active VSG ES (35 lac operator repeats; arrowheads). (E–M) 2K1N bloodstream form cell lines tagged with GFP in the rDNA locus (n = 170), inactive VSG ES (n = 201), or active ES (n = 186) were grouped based on DAPI staining (Fig. S2) as G2 prophase, metaphase, or anaphase and scored as containing either one or two GFP dots. Representative pictures show GFP dot detection (arrowheads) in cell lines GFP tagged in the rDNA locus (E–G), inactive VSG ES (H–J), and active VSG ES (K–M). Insets show DAPI signal distribution by applying a gradient LUT (see Materials and methods), for which red corresponds to lower intensity pixel values and green to higher values. (C–M) Asterisks mark the kinetoplast. Bars, 1 µm.
To confirm the delay in active VSG ES sister chromatid separation compared with other loci, we generated a cell line that enabled us to simultaneously visualize the active VSG ES and rDNA loci in the same nucleus. For this purpose, we generated a double GFP chromosome–tagged bloodstream cell line in which 35 lac operator repeats were inserted in the active VSG ES, whereas 250 lac operator repeats were inserted in the rDNA locus. Upon IF with anti-GFP antibodies and DAPI staining, the difference in the number of lac operator repeats was manifest in a clear difference in GFP dot intensity, making it possible to discriminate between the two loci in a single nucleus (Fig. 2 C). Analysis of 2K1N cells suggests that the separation of sister chromatids of the rDNA locus precedes the separation of sister chromatids of the active VSG ES (Fig. 2 D).
Sister chromatids of the active VSG ES remain associated to the single ESB until chromosome segregation
To determine whether the observed decrease in sister chromatid separation of the active VSG ES reflects a delay in chromatid release during mitosis, we analyzed the separation of sister chromatids during mitotic progression in the 2K1N cell population. Although trypanosomes do not visibly condense their chromosomes during mitosis, changes in DNA distribution during mitotic phases are evident (Ersfeld and Gull, 1997; Ogbadoyi et al., 2000; Kumar and Wang, 2005). Thus, a gradient red/green look up table (LUT) applied to the DAPI signal (red for lower intensity pixel values and green for higher values) facilitates the grouping of cells into three DNA distributions: homogenous (late G2–early mitosis; Fig. S2 A), concentration of the bulk of DNA in the nuclear center (metaphase like; Fig. S2 B), and two stretched and separating nuclei (anaphase like; Fig. S2 C). Determination of sister chromatid separation showed that sisters of the rDNA locus and the inactive VSG ES are clearly separated throughout mitosis (Fig. 2, E–J). In contrast, separation of sister chromatids of the active VSG ES was only detected during chromosome segregation in anaphase-like cells (95% of the cells showing two GFP dots; Fig. 2, K–M), demonstrating that the release of sisters of the active VSG ES is delayed compared with other loci studied, including a VSG ES in the inactive transcriptional state (Fig. 2, H–J).
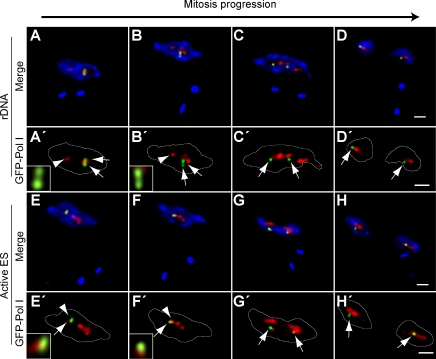
The observed delay in the separation of sister chromatids of the active VSG ES could be explained by their association with the same and unique ESB present in the nucleus until chromosome segregation. To test this hypothesis, we analyzed the localization of sisters of the active VSG ES and rDNA loci with respect to pol I machinery during mitosis. Early during mitosis, sister chromatids of the rDNA locus can be found separated but in association with the bulk of pol I machinery corresponding to the nucleolus (Fig. 3, A–B′). Upon mitosis progression, sister chromatids of the rDNA locus are segregated to opposite poles of the nucleus in association with pol I machinery, rendering undistinguishable pol I bodies (Fig. 3, C–D′). In contrast, sisters of the active VSG ES remain held together and in association with the unique ESB present in the nucleus (Fig. 3, E–F′) until chromosome segregation, when they appear separated but still in association with pol I machinery (presumably all or part of the ESB), giving rise to two daughter nuclei each containing a replicated copy of this locus (Fig. 3, G–H′).
Figure 3.
Sisters of the active VSG ES remain associated to the unique ESB until chromosome segregation. Different cell lines with a GFP tag in either the rDNA locus or the active VSG ES were subjected to double indirect 3D IF using anti-TbRPA1 antiserum, monoclonal anti-GFP antibodies, and DAPI staining. The positions of GFP dots and pol I machinery were determined during different mitotic phases as defined by DAPI staining (Fig. S2). (A–H) Maximum intensity projections of three-channel 3D representative stacks showing anti-TbRPA1 signal in red, anti-GFP signal in green, and DAPI staining in blue in the cell line GFP tagged in the rDNA (A–D) or active VSG ES (E–H). (A′–H′) Maximum intensity projections of the anti-TbRPA1 and anti-GFP signals. Insets show single planes of anti-TbRPA1 and anti-GFP signals for the slice in which the maximum GFP dot intensity was detected. The yellow color indicates colocalization. White lines indicate the nuclear outline. Arrows indicate the GFP dot signal, whereas arrowheads mark the position of the ESB. Bars, 1 µm.
TbSCC1 is required for regulation of sister chromatid separation of the active VSG ES
Given that the delay in VSG ES chromatid separation is only detected when the VSG ES is in an active transcriptional state (Fig. 2), we postulated that sister chromatid cohesion could be important for the correct inheritance of VSG monoallelic expression. Cohesins are highly conserved proteins involved in holding sister chromatids together in eukaryotes (Nasmyth, 2002). Thus, we decided to analyze whether disruption of the cohesin complex results in premature separation of sisters of the active VSG ES. To accomplish this, we performed a conditional knockdown using a tetracycline-inducible RNAi system (Ngo et al., 1998) for the orthologue of the yeast cohesin subunit gene Scc1 in T. brucei (TbSCC1; Gluenz et al., 2008; Sharma et al., 2008) in GFP chromosome–tagged cell lines for the rDNA locus and the active VSG ES.
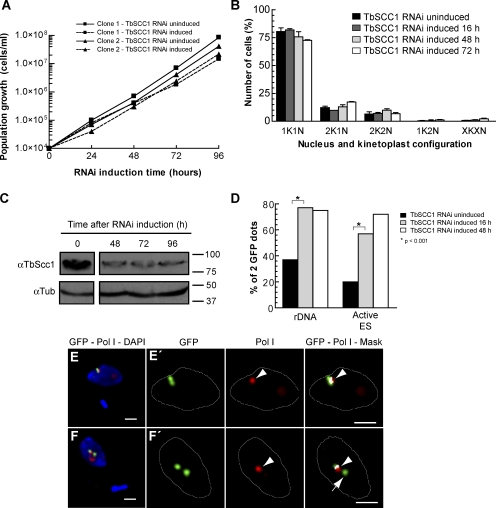
Similar to what occurs in other eukaryotes (Uhlmann, 2001a; Nasmyth, 2002), a profound depletion of TbSCC1 by RNAi is known to lead to a cell division phenotype in bloodstream trypanosomes (Gluenz et al., 2008). However, partial depletion of SCC1 levels has been shown not to compromise cell division progress, making the analysis of secondary roles of the cohesin complex possible, even during the developmental processes of multicellular organisms (Rollins et al., 2004; Dorsett, 2007; Horsfield et al., 2007). To avoid indirect effects caused by the malfunctioning of mitosis, we chose cell lines in which the induction of TbSCC1 knockdown did not result in a decreased growth rate (Fig. 4 A) nor arrest of cell division (Fig. 4 B). Western blot analysis of cell extracts using anti-TbSCC1 antiserum confirmed that although induction of TbSCC1 RNAi resulted in an evident depletion of the target protein, cells still expressed a significant amount of TbSCC1 during the first 4 d of RNAi induction (Fig. 4 C).
Figure 4.
Knockdown of TbSCC1 results in a decrease in sister chromatid cohesion in the active VSG ES and loss of association with the ESB. Bloodstream form cell lines that contain a tetracycline-inducible partial knockdown of the cohesin subunit TbSCC1 were selected and characterized. (A) Growth curves of two independent clones in which RNAi of TbSCC1 has been induced (doxycycline+) compared with uninduced cultures (doxycycline−). (B) Cell cycle progression as revealed by quantification of nucleus and kinetoplast configurations after different induction times of TbSCC1 knockdown. Mean ± SD of the percentages (n > 300 for each time point) obtained for two independent clones are shown. (C) Western blot using anti-TbSCC1 antiserum (Sharma et al., 2008) showing a partial depletion of the levels of the TbSCC1 protein after different periods of induction of TbSCC1 RNAi. Molecular mass is indicated in kilodaltons. (D) GFP-tagged cells lines were subject to indirect IF using anti-GFP antibodies as well as DAPI staining and were recorded as displaying one or two GFP dots in 2K1N cells. The histogram shows the percentages of cells displaying two GFP dots corresponding to duplicated and separated sister chromatids of either the active VSG ES or the rDNA locus at different time points after induction of TbSCC1 knockdown (rDNA: uninduced [n = 115], induced 16 h [n = 110], and induced 48 h [n = 103]; active VSG ES: uninduced [n = 112], induced 16 h [n = 107], and induced 48 h [n = 104]). (E and F) A cell line with a GFP tag in the active VSG ES was subjected to double indirect 3D IF using anti-TbRPA1 antiserum, monoclonal anti-GFP antibodies, and DAPI staining upon 4 d of induction of TbSCC1 down-regulation. Maximum intensity projections of three-channel 3D representative stacks showing anti-TbRPA1 signal in red, anti-GFP signal in green, and DAPI staining in blue in 1K1N cells. (E′ and F′) Colocalization analysis of the GFP dot signals with respect to the ESB. Maximum intensity projections of the subset of slices containing GFP dot signal are shown. A colocalization mask (white) was calculated for each nonequalized 8-byte slice and merged with both anti-GFP (green) and anti-TbRPA1 (red) labeling. Arrowheads indicate the position of the ESB, whereas the arrow marks a sister chromatid of the active VSG ES that is no longer associated with the ESB. White lines indicate the nuclear outline. Bars, 1 µm.
To investigate whether TbSCC1 has a role in the delay of sister chromatid separation in the active VSG ES, we performed indirect IF using anti-GFP antibodies and DAPI staining upon partial depletion of TbSCC1 in cell lines with either active VSG ES or rDNA sequences tagged with GFP. Consistent with the role of the cohesin complex in holding sister chromatids together in other organisms, upon depletion of TbSCC1 for 16 h, sister chromatids of the rDNA locus were separated in a higher percentage of cells compared with the uninduced culture (Fig. 4 D). Similarly, after 16 h of TbSCC1 depletion, sister chromatids of the active VSG ES were separated in an abnormally high proportion in 2K1N cells (57%; Fig. 4 D). This percentage increased to 72% upon 48 h of induction, which is similar to that for the rDNA locus (75%; Fig. 4 D), showing that the partial depletion of TbSCC1 results in the separation of sister chromatids of both active VSG ES and rDNA loci to the same extent. Similar results were obtained upon RNAi with a nonoverlapping fragment of TbSCC1 (unpublished data).
Next, we addressed whether induced early separation of sister chromatids of the active VSG ES has an effect on the association of both sisters with the single ESB. According to the ESB single-locus occupancy model for VSG ES monoallelic expression (Navarro and Gull, 2001), one would predict that upon cohesin depletion and premature sister separation, one of the sisters might no longer be associated with the ESB. We performed 3D IF using anti-TbRPA1 and anti-GFP antibodies on the active VSG ES GFP-tagged cell line upon TbSCC1 depletion by RNAi. After 2 d of partial depletion of TbSCC1, sister chromatids of the active VSG ES were detected separated from one another in G2 cells. Among these cells, 45.4% showed that slightly separated (GFP doublet) sister chromatids of the active VSG ES remain associated with the single ESB present in the nucleus (Fig. 4, E and E′). Importantly, in 26.7% of the cells displaying two GFP dots, sister chromatids of the active VSG ES were clearly separated, and one of them was no longer associated with the ESB (Fig. 4, F and F′), suggesting that cohesion between sister chromatids of the active VSG ES is important in maintaining their stable association with the unique transcriptional ESB.
Cohesin complex integrity is required for the correct regulation of VSG monoallelic expression
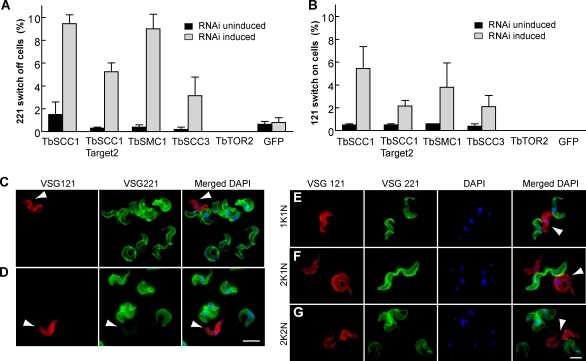
TbSCC1 depletion resulted in an early separation of sister chromatids of the active VSG ES and a reduced association with the ESB (Fig. 4), suggesting that cohesins might play a role in the regulation of VSG monoallelic expression. To test this hypothesis, we first analyzed the frequency of VSG switching off in cells that were partially devoid of TbSCC1. IF analysis of TbSCC1-depleted trypanosomes using specific anti-VSG221 antibodies showed a percentage of cells that switched their surface coat from VSG221 to a different VSG that was significantly higher (9.5 ± 0.5%) than the noninduced culture (1.3 ± 0.7%; Fig. 5 A). As controls, depletion of either an essential protein kinase (TbTOR2; Barquilla et al., 2008) or the GFP protein did not result in an increase in VSG221-negative cells (Fig. 5 B). Moreover, depletion of various essential proteins such as TbActin, TbMyosin 1B, and TbRPB5 by RNAi (Fig. S3 A) did not result in changes in the switching off frequency either (Fig. S3 B). Deleterious phenotypes could lead to VSG shedding from the cell surface, which can be detected in the culture supernatant. However, TbSCC1 depletion did not yield detectable soluble VSG, whereas TbActin and TbMyosin depletion by RNAi did (Fig. S3 C). These data show that possible indirect effects related to cell viability or overload of the RNAi machinery are insufficient to cause an increase in the frequency of VSG221 switching off.
Figure 5.
Knockdown of TbSCC1 results in an increase in the in situ switching to VSG121 ES. (A) RNAi-mediated knockdown of different subunits of the cohesin complex was induced over 4 d in bloodstream form trypanosomes. Indirect IF using anti-VSG221 or anti-VSG121 antibodies and DAPI staining was performed. (A and B) The frequency of cells negative for VSG221 labeling (A) or positive for VSG121 labeling (B) is shown. Mean ± SEM of experiments (n > 500) for two independent clones is shown for each gene. (C and D) Double indirect IF with rabbit anti-VSG221 (green), mouse anti-VSG121 (red), and DAPI staining (blue) was performed upon TbSCC1 knockdown. Representative pictures show a trypanosome positive only for VSG121 labeling (C; arrowheads) or positive for both VSGs (D; arrowheads). Yellow indicates double labeling. (E–G) Knockdown of TbSCC1 promotes the appearance of VSG121-expressing variants that are able to proceed through the cell cycle. Representative pictures showing bloodstream trypanosomes positive only for VSG121 labeling and displaying different nucleus/kinetoplast numbers are shown: 1K1N (E), 2K1N (F), and 2K2N (G). Arrowheads indicate the position of the cell in which expression of the VSG gene has changed from VSG221 to VSG121. Bars, 5 µm.
Antigenic variation in T. brucei can occur by either recombination of a new vsg gene into an already active VSG ES telomere or by transcriptional activation of another VSG ES telomere and concomitant inactivation of the previously active one, known as an in situ switch (Pays et al., 2004). To analyze which switching type is induced by TbSCC1 depletion, we performed IF analysis using anti-VSG antibodies that specifically recognize VSG121, which is codified by a VSG gene located in an inactive VSG ES telomere and activated mainly by in situ switching in this cell line (Aitcheson et al., 2005). Thus, the number of VSG121-positive cells can be used as an indirect measurement of the in situ switching on frequency of inactive VSG ES telomeres. A 10-fold increase in the number of cells expressing VSG121 was detected after 4 d of induction of TbSCC1 knockdown, from 0.5 ± 0.1% to 5.4 ± 1.3% (Fig. 5 B), suggesting that TbSCC1 is required for proper inheritance of the active VSG ES transcriptional state. No increase in the activation of the inactive VSG121 ES was detected upon depletion of control proteins such as TbTOR2, GFP, TbActin, TbMyosin 1B, and TbRPB5 (Fig. 5 B and Fig. S3 B). To dismiss a possible RNAi off-target effect, we performed the same analysis using a nonoverlapping fragment of the TbSCC1 gene as the target sequence. A similar result to the initial construct illustrated the specificity of TbSCC1 knockdown in promoting an increase in in situ VSG switching (Fig. 5, A and B). Likewise, an increase in the switching frequency was also detected when other subunits of the cohesin complex such as TbSMC1 or TbSCC3 were depleted by RNAi (Fig. 5, A and B), indicating that the observed switching phenotype is a consequence of compromising the cohesin complex rather than a cohesin-independent function of TbSCC1.
Next, we analyzed surface VSGs in the new VSG121 variants by double IF with rabbit anti-VSG221 antiserum and a mouse monoclonal anti-VSG121 antibody upon 4 d of TbSCC1 partial depletion. Among the cells positive for VSG121, a considerable percentage did not display VSG221 on their surface (19.6%; Fig. 5 C), showing that VSG121-expressing trypanosomes do not arise as the result of simultaneous coactivation of both VSG221 ES and VSG121 ES. However, we also detected a high proportion of trypanosomes (80.4%) that highly express VSG121 but were still faintly positive for VSG221 labeling (Fig. 5 D); these most likely represent trypanosomes in which the change of the coat from VSG221 to VSG121 has not yet finished because of the long half life (32 h) of the VSG protein (Bulow et al., 1989). New variants expressing exclusively VSG121 displayed all three nucleus–kinetoplast configurations, showing that they are viable cells able to correctly progress through the cell cycle (Fig. 5, E–G).
To avoid VSG121-expressing trypanosomes at the time of TbSCC1 RNAi induction (0.5%), we recloned and analyzed the trypanosomes arising directly from a single VSG221-positive cell to estimate the switching frequency to VSG121 upon TbSCC1 depletion. Antigenic variants were analyzed by FACS in two independent TbSCC1 RNAi subclones. Cultures were split into uninduced and induced TbSCC1 RNAi and double labeled with anti-VSG221 and anti-VSG121 antibodies. FACS analysis showed 0.03 and 0.01% VSG121-positive cells in two uninduced clones (Fig. S4 A), which is similar to previous results (Aitcheson et al., 2005). However, upon TbSCC1 depletion, the percentage of VSG121-positive cells among these clones rose from 0.03 to 0.3% and from 0.01 to 0.16%, representing a 10-fold increase in the in situ activation of the VSG121 ES. In addition, a 20-fold increase in the frequency of VSG221 ES switching off was also detected by FACS (Fig. S4, A and B). A similar increase in VSG221-negative and VSG121-positive cells was observed when TbSCC1 RNAi was performed on a cell line containing a resistance gene downstream of the promoter in the active VSG221 ES (Penate et al., 2009), which allows selection to keep the VSG221 ES active before RNAi induction (Fig. S4 C).
Partial depletion of TbSCC1 induces in situ transcriptional switching to different telomeric VSG ESs
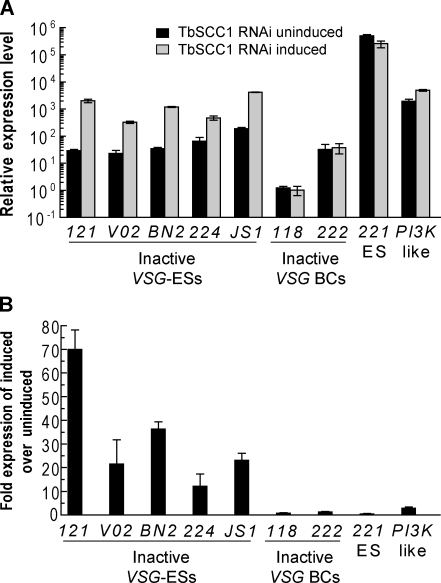
A recent study has provided the sequences of 15 telomeric VSG ESs in the T. brucei Lister 427 strain used in this study (Hertz-Fowler et al., 2008). Thus, we can address whether depletion of the cohesin complex also results in the in situ activation of other previously inactive telomeric VSG ESs. To do so, we performed quantitative RT-PCR (qRT-PCR) analysis using specific primers for different inactive telomeric VSG genes upon 4 d of TbSCC1 depletion (Fig. 6 A). Interestingly, we detected a significant increase in the mRNA expression levels of not only VSG121 (69 ± 8–fold) but also of all of the other inactive telomeric VSG genes tested such as VSGVO2 (21 ± 10–fold), VSGBn2 (36 ± 3–fold), VSG224 (12 ± 5–fold), and VSGJS1 (23 ± 2.9–fold), whereas the housekeeping gene PI3K-like was not significantly affected (Fig. 6 B). In agreement with the detected increase in the active VSG221 ES switching-off rate by IF (Fig. 5 A), a corresponding reduction in the VSG221 mRNA level of 51 ± 13% was also detected by qRT-PCR (Fig. 6 A). To confirm that our RT-PCR analysis reflects real antigenic switches, we performed IF analysis using anti-VSGVO2 antibodies and found that cohesin depletion increases the frequency of in situ activation of independent telomeric VSG genes such as VSGVO2 (Fig. S5).
Figure 6.
Knockdown of TbSCC1 results in the in situ transcriptional activation of inactive telomeric VSG ESs. (A) Histogram showing the relative expression of mRNA measured by qRT-PCR of VSG genes located in different VSG ESs (ES) or different promoter-lacking VSG BC genes upon 4 d of induction of TbSCC1 RNAi. An increase in the expression of VSG genes located in previously inactive VSG ESs is detected, whereas VSG-BCs remain at a similar level when compared with the uninduced control. Expression values are plotted in relative expression units, calculated relative to myosin expression, and multiplied by 1,000 for better visualization on a logarithmic scale. (B) Histogram showing the fold change in gene expression levels (TbSCC1 RNAi induced/TbSCC1 RNAi uninduced) shown in A. (A and B) Mean ± SD of three different measures is shown.
In addition, we wanted to investigate whether switching by recombination of VSG genes into the active telomeric VSG ES was also induced upon TbSCC1 knockdown. There are hundreds of BCs of VSG genes (VSG-BC) located in internal chromosomal arrays with no associated promoters, which can only be transcribed when the gene is duplicated by recombination and inserted into the active VSG ES. qRT-PCR showed no significant changes in the mRNA levels of either of the two VSG BC genes tested (Fig. 6 A), suggesting that the VSG recombination rate was not affected upon knockdown of TbSCC1. Together, these results suggest that the increase in the switching-off rate of the active VSG221 ES corresponds with the random activation of other, previously inactive VSG ESs.
Discussion
Monoallelic expression in multigene families is a poorly understood phenomenon in a variety of model systems. After the identification of the association of the ESB with the active VSG ES, it has been suggested that such a structure could mediate the monoallelic expression of a single VSG gene through a mechanism that involves the stable association with just 1 out of ∼15 VSG ESs (Navarro and Gull 2001; Navarro et al., 2007). Furthermore, this model could apply more widely to the regulation of monoallelic expression in other multiallelic gene families (Borst, 2002).
The active VSG ES is selectively targeted for delayed separation of sister chromatids
The release of sister chromatids of the active VSG ES is delayed until chromosome segregation, which is in contrast to chromatid separation early during mitosis of all other loci studied (Fig. 2). Interestingly, enhanced cohesion of the VSG ES seems to be selectively coupled to its monoallelic transcriptional state, as the same enhanced cohesion is not observed in the inactive VSG ES nor in the procyclin gene family, suggesting that it is a unique feature of the fully active and monoallelically expressed VSG ES rather than simply related to pol I transcription of protein-coding genes. Importantly, we show that enhanced cohesion of sister chromatids of the active VSG ES depends on the cohesin complex, as induction of TbSCC1 depletion prompted premature separation of sister chromatids of the active VSG ES.
Compromising the integrity of the cohesin complex also resulted in early separation of sister chromatids of the rDNA locus. However, this does not imply that the cohesin complexes that target both loci are identical, as existing data suggest that different populations of cohesin complexes target different genomic loci that are subjected to differential regulation and carry out distinct functions (for reviews see Guacci, 2007; Losada, 2007). In metazoan organisms, the unloading of different populations of cohesin complexes from different chromosomal regions relies on a biphasic kinetic of chromosome separation (for review see Guacci, 2007). We observe that similar biphasic kinetics also occur between the active VSG ES and control loci in trypanosomes, suggesting that this delay in sister chromatid separation may be caused by the differential unloading of a distinct population of cohesin complexes present in the active VSG ES.
Association of active VSG ES sister chromatids with the ESB is maintained by the cohesin complex
We show that the single ESB is not duplicated upon genome duplication, but rather, a delay in the separation of sister chromatids of the active VSG ES facilitates the association of both sisters with the single ESB present in the nucleus until chromosome segregation (Figs. 1 and 2). This delay in separation of sisters may serve as a mechanism to assure that newly replicated VSG ES chromatids interact with similar nuclear machinery such as the ESB or particular chromatin modifiers potentially enriched in the ESB throughout the entire cell cycle. During mitosis, the trypanosome nucleolus does not disintegrate but rather splits into two clusters of similar size preceding karyokinesis (Ogbadoyi et al., 2000). A similar redistribution of the pol I–containing ESB material in association with the active VSG ES into daughter nuclei upon chromosome segregation may be key to maintain the VSG ES active transcriptional state in the next generation.
Upon TbSCC1 knockdown, the delay that we originally observed in the separation of sister chromatids of the active VSG ES was abolished. It is important to note that we studied trypanosomes in which the decrease of TbSCC1 protein level upon RNAi was only partial, to avoid previously described cell cycle defects (Gluenz et al., 2008). Using 3D IF analysis, we found that upon induced separation of sister chromatids of the active VSG ES, one of the sisters was no longer associated with the ESB in a considerable percentage of cells (Fig. 4). Interestingly, this result raises the possibility that the transcriptional inactivation of the active VSG ES upon TbSCC1 knockdown is a consequence of the loss of association of sister chromatids of this locus with the ESB. We cannot rule out the possibility that the lack of association with the ESB is an indirect result of transcriptional inactivation of the active VSG ES; however, one of the sisters still remained associated with the ESB (Fig. 4 F), suggesting this is not the case. Alternatively, cohesin may be required to maintain the active transcriptional state of the replicated VSG ES through the loading of chromatin remodeling and insulator factors. In any case, these results suggest that the integrity of the cohesin complex is required to maintain the normal association between sisters of the active VSG ES and the ESB until chromosome segregation.
Inheritance of the active transcriptional state of the VSG ES requires the cohesin complex
We show that RNAi-mediated depletion of three different subunits of the cohesin complex (TbSCC1, TbSMC1, and TbSCC3) results in the appearance of cells that are negative for VSG221 and the concomitant appearance of cells positive for VSG121. Double IF experiments with anti-VSG221 and anti-VSG121 antibodies show that after TbSCC1 knockdown, 19.6% of VSG121-expressing trypanosomes do not exhibit VSG221 in their surface, indicating that the VSG221 ES has been switched off. This result rules out the possibility that both VSG ESs are fully activated at the same time upon TbSCC1 depletion in these cells. The detection by IF of VSG121 on the surface is an indirect measure of in situ activation of the telomeric VSG121 ES (Aitcheson et al., 2005). Furthermore, we detected additional telomeric VSG ES activation by detecting VSGVO2 on the surface of independent switchers. Gene expression analysis shows that TbSCC1 knockdown results in the transcriptional activation of several VSG genes located in additional telomeric VSG ESs such as VSGBn2 ES, VSG224 ES, and VSGJS1 ES (Fig. 6). These data suggest that compromising the integrity of the cohesin complex in T. brucei prompts independent in situ transcriptional switches to several previously inactive VSG ESs.
The number of switch-off variants generated within only 4 d of partial depletion was clearly significant with respect to noninduced cultures, albeit usually no higher than ∼10% (Fig. 5 A). Although this number may appear small, given the low frequency of in situ switching in this cell line and the time frame in which we have performed our experiments, it represents a surprising increase in the switching rate upon a protein depletion that has not been previously described. Cohesin depletion results in a 10% switching-off rate of cells expressing VSG221 on their surface; however, IF analysis of the cells positive for switching on of VSG121 showed that 79.4% of them still express the former VSG221. This suggests that we may be underestimating the true number of cells undergoing VSG221 switching off. Although we lack antibodies against every possible newly expressed VSG, RT-PCR analysis suggests that telomeric VSG genes become active in a variable proportion of cells upon cohesin subunit knockdown and detection of independent VSG variants expressing previously inactive telomeric VSG genes on the surface such as VSGVO2 confirmed this hypothesis (Fig. S5). Altogether, these data suggest that the cohesin complex is required for the maintenance of the VSG ES active state, and impairing cohesin function results in an increase in VSG switching frequency.
Several observations suggest that cohesins play a role in the regulation of gene expression. In zebrafish, the cohesin complex has been implicated in such regulation (Dorsett, 2007; Horsfield et al., 2007), whereas in Saccharomyces cerevisiae, cohesins are involved in Sir-mediated silencing (for review see Losada, 2007). In mammals, cohesins have been recently shown to regulate the insulin-like growth factor 2 locus (Wendt et al., 2008). Although molecular mechanisms remain largely unknown, data to date suggest that the cohesin complex can act as a transcriptional insulator protein (for review see Peric-Hupkes and van Steensel, 2008). Cohesins are also known to be loaded onto long repeat elements within constitutive heterochromatin (for review see Losada, 2007). Interestingly, clear boundaries for transcriptional regulation in the VSG ES include nontranscribed repeat sequences such as the 50-bp repeats expanding tens of kilobases upstream of the VSG ES promoter (Sheader et al., 2003) as well as the telomeric sequences downstream of the VSG gene.
In mammals, cohesin complexes are targeted to specific sites on chromosomes and function in the regulation of gene expression (for review see Peric-Hupkes and van Steensel, 2008). Recent data show an interplay between transcriptional insulators and cohesins function (Parelho et al., 2008; Wendt et al., 2008). As a consequence, insulator-mediated enrichment of cohesin complexes is sensitive to different epigenetic marks. A similar DNA-binding protein operating in T. brucei, capable of integrating both sequence and epigenetic status, would allow the targeting of cohesin to the active VSG ES, providing a mechanism to integrate different epigenetic mechanisms to ensure monoallelic expression. Removal of cohesins could then hamper the proper insulation of the active VSG ES and result in its transcriptional inactivation.
Another interesting mechanistic link emerges in the relationship between cohesin and chromatin remodeling factors (Hakimi et al., 2002). In T. brucei, it has been recently reported that RNAi of a member of the ISWI family derepressed silenced chromatin (Hughes et al., 2007). Recent studies showed that the histone methyltransferase TbDOT1 and the telomeric-associated protein TbRAP1 are required to maintain strict VSG ES silencing (Figueiredo et al., 2008; Yang et al., 2009). However, depletion of these proteins did not result in VSG ES in situ switching but rather an increase in transcription of silenced telomeric VSG ESs (Figueiredo et al., 2008; Yang et al., 2009).
Data to date suggest that important similarities govern monoallelic expression in T. brucei (Figueiredo et al., 2008; Yang et al., 2009) and Plasmodium falciparum (Ralph and Scherf, 2005; Lopez-Rubio et al., 2007), where chromatin modification is required to maintain strict telomeric gene silencing. Cohesins are required to prevent spreading of heterochromatin from silenced loci (for review see Losada, 2007), suggesting that upon cohesin knockdown, silenced chromatin in the VSG ES telomeres may also be affected, facilitating the transcriptional switches we detected.
Several models have been suggested to explain monoallelic expression in a variety of experimental systems from protozoa to mammals. DNA–DNA interactions seem to be involved in olfactory gland receptor expression, where monoallelic expression by olfactory neurons may occur via the interaction between an enhancer element in trans and a single promoter (Lomvardas et al., 2006; Fuss et al., 2007) Importantly, although the role of the cohesin complex has not yet been explored in mammalian multiallelic families, it has been linked to DNA–DNA interactions and to chromatin insulator proteins that regulate enhancer–promoter interactions (for reviews see Losada, 2007; Gondor and Ohlsson, 2008; Peric-Hupkes and van Steensel, 2008). Although, in trypanosomes, no such enhancer has yet been described, the effect of cohesin depletion on VSG ES monoallelic expression may suggest the alteration of a putative DNA–DNA interaction between a hypothetical enhancer element and the VSG ES promoter. It is tempting to speculate that cohesins may play a role in the molecular mechanism underlying monoallelic expression throughout multiallelic families, and certainly, this concept warrants further investigation.
Collectively, our results provide a new model for how the active transcriptional state of a monoallelically expressed locus is inherited through a cohesin-mediated mechanism. In addition to ensuring sister chromatid cohesion, cohesin function in this divergent eukaryote is required to prolong the association of active VSG ES sister chromatids both with each other and with the unique ESB. Upon cohesin depletion, premature sister chromatid separation uncouples the association of one sister with the ESB, supporting the ESB single occupancy model for VSG ES monoallelic expression. These data provide evidence for a novel role of the cohesin complex in the epigenetic regulation of monoallelic expression of the VSG ES in T. brucei.
Materials and methods
Trypanosomes
Throughout this study, T. brucei bloodstream (Molteno Institute Trypanozoon antigenic type [MITat] 1.2; clone 221a) and 427 procyclic forms were used. DNA transfection and selection procedures have been described previously (Wirtz et al., 1999).
GFP-LacI repressor tagging of chromosomes
Single GFP-tagged locus cell lines used in this study have been described previously (Navarro and Gull, 2001; Landeira and Navarro, 2007). As initial cell lines, T. brucei bloodstream form single marker (Wirtz et al., 1999) and procyclic form 1,313–1,333 were used. After electroporation with a pLew100 (Wirtz et al., 1999) derivate containing the eGFP-LacI-SV40nls gene fusion (Navarro and Gull, 2001), stable transformed clonal cell lines (Landeira and Navarro, 2007) that express the GFP-LacI fusion in a tetracycline-dependent manner were obtained. 250 lac operator repeats were stably inserted in the locus of interest by homologous recombination. lac operator repeat insertion plasmids for the active VSG221 ES, inactive VSG121 ES (Navarro and Gull, 2001), rDNA, VSG121-BC, and procyclin (GPEET/PAG3) were constructed containing a variable target DNA, a 256 lac operator repeat sequence, and the promoter of the locus under study to drive the selectable marker, as described previously (Landeira and Navarro, 2007). The double GFP-tagged cell line was obtained by the stable transformation of the rDNA GFP–tagged cell line (Landeira and Navarro, 2007) with a plasmid targeting the active VSG221 ES containing 35 lac operator repeats and a hygromycin selectable marker, a derivate construct of the one described previously (Navarro and Gull, 2001). GFP-LacI expression was induced in early exponentially growing cultures with 1 µg/ml doxycycline (Sigma-Aldrich) for 16 h.
YFP-TbRPB5z–expressing trypanosomes
The TbRPB5z gene was cloned by PCR using oligonucleotides 5′-GGTGGCAGGCCGGCCATGGATTCCGTCAAAAGCTT-3′ and 5′-AGAGGTGGCCGGCCTAGCTAAATAACATGTCGAT-3′, digested with FseI, and cloned into pTAP-YFP, resulting in a YFP-TbRPB5z open reading frame under the control of an tetracycline-inducible procyclin promoter. Bloodstream form single marker trypanosomes were transfected and selected using procedures previously described (Wirtz et al., 1999).
RNAi experiments
The TbSCC1 RNAi construct was made using the vector p2T7Bla to allow tetracycline-inducible expression of double-stranded RNA from a T7 promoter in bloodstream form cell lines GFP tagged in either the active VSG ES or in the rDNA locus. A fragment corresponding to ∼500 nucleotides of different open reading frames was amplified by PCR with the following oligonucleotides: TbSCC1, 5′-GCGGATCCAATGGGTCGCACGAGGAAGC-3′ and 5′-GCAAGCTTCGTCACCGTCATCAGGAAGC-3′; TbSCC1 fragment 2, 5′-GCGGATCCCGGCTCTCACAGCCTCTTAC-3′ and 5′-GCAAGCTTGTAAGGCCAAGCTCCACATC-3′; TbSMC1, 5′-GCGGATCCTCTCAGTTTCAGCGAACCCT-3′ and 5′-GCAAGCTTAGGTTCAAACACGCGATACC-3′; TbSCC3, 5′-GCGGATCCTTCTCGTGTGGCTTCTGATG-3′ and 5′-GCAAGCTTACAAACGAATTTCAGGTGCC-3′; and TbTOR2 oligonucleotides were previously described (Barquilla et al., 2008). PCR fragments were cloned into the BamHI and HindIII sites of p2T7Bla. RNAi stable cell lines were diluted daily so that the density of the cultures was maintained between 105 and 106 in media without selection drugs. Double-stranded RNA synthesis was induced by the addition of 1 µg/ml of the tetracycline analogue doxycycline in every dilution step. Analysis of VSG switching frequency upon protein depletion was performed using recently obtained clones. Western blotting was performed using standard procedures with affinity-purified polyclonal antisera against TbSCC1 raised in rabbit (Sharma et al., 2008).
IF
2D IF fixation was performed with 3% paraformaldehyde for 20 min. Permeabilization with 1% NP-40 (GFP-LacI IF) or 0.1% Triton X-100 (VSG IF) for 20 min was followed by incubation in 0.5% blocking reagent (Roche) for 20 min. Rabbit polyclonal anti-GFP (1:5,000; Invitrogen), anti-VSG221, or anti-VSG121 antibodies diluted in 0.5% blocking reagent were used as a primary antibody. Goat anti–rabbit Alexa Fluor 488–conjugated antibodies were used as a secondary antibody. ProLong Gold antifade with DAPI reagent (Invitrogen) was used as a mounting solution. 3D IF in trypanosomes (Engstler and Boshart, 2004) of GFP-tagged cell lines was performed as described previously (Landeira and Navarro, 2007). Cells were fixed in 4% paraformaldehyde on ice for 2.5 h and permeabilized in 1% NP-40 at room temperature for 1 h. Monoclonal anti-GFP (1:300; Invitrogen) and affinity-purified antiserum against the large subunit of RNA polymerase I (anti-TbRPA1; 1:600; Navarro and Gull, 2001) were diluted in 1% blocking reagent (Roche). Monoclonal anti-BB (BBA4; Woodward and Gull, 1990) supernatant was used undiluted. Alexa Fluor 488– or Alexa Fluor 594–conjugated goat species–specific antibodies (Invitrogen) were used as a secondary antibody. Cells were DAPI stained and mounted on slides. Stack (0.1-µm z step) acquisition was performed with a motorized microscope system (Cell R IX81; Olympus) equipped with a 100× NA 1.4 oil objective (Olympus), MT20 illumination system, and a charge-coupled device camera (Orca; Hamamatsu Photonics). Deconvolution of 3D datasets was performed as described previously using Huygens Essential version 2.9 software (Scientific Volume Imaging B.V.; Engstler and Boshart, 2004). Point spread function was measured using 0.2-µm tetraspeck microspheres (Invitrogen). Colocalization analysis, pseudocoloring, gradient LUTs, and maximum intensity projections were performed using ImageJ version 1.37 software (National Institutes of Health).
Statistical analysis
For each GFP-tagged cell line, GFP-LacI–expressing cells where analyzed, and single spots were defined as signals with a uniform spherical appearance. Scoring was performed by direct optical observation, and questionable cells were analyzed by 2D or 3D digital imaging. Whenever percentages of different cell lines were averaged, Student's t test was applied. Percentages obtained for a particular cell line were compared between the indicated categories using Fisher's exact test. Statistical significance was determined by using a 95% confidence interval.
Purification of VSG121
VSG121 was purified according to a previously described protocol (Cross, 1984) from a T. brucei cell line expressing VSG121 (Navarro and Cross, 1996). In brief, release of VSG from glycophosphatidylinositol anchors was obtained after osmotic lysis at 0°C for 15 min. After centrifugation, the membranes present in the pellet were incubated for 5 min at 37°C in distilled water with 0.5 mM zinc acetate, allowing for the liberation of surface proteins. VSG121 was collected in the supernatant after a last centrifugation. Purity of the proteins was checked by Coomassie-stained SDS-PAGE.
Anti-VSG121 monoclonal antibody
A BALB/c mouse was immunized by three intraperitoneal injections of 100 µg of purified VSG121. 3 d before fusion, 40 µg of the protein was injected intravenously. Normal antibody-producing spleen cells were fused with immortal myeloma cells. The hybridoma that produced the desired antibody was identified by ELISA, Western blotting, and IF and subcloned to obtain a single epitope antibody, which was named V7B11F2. This antibody was tested against VSG221-expressing cells by Western blotting and IF to ensure a lack of cross-reactivity.
FACS assays
After washing in PBS, ∼2 × 107 cells were fixed in 1% paraformaldehyde overnight at 4°C. Cells were then washed and labeled with rabbit polyclonal anti-VSG221 (1:1,000) and mouse monoclonal anti-VSG121 (1:500) antibodies as a primary antibody. Alexa Fluor 488– and Alexa Fluor 594–conjugated goat species–specific antibodies were used as a secondary antibody. Labeled cells were analyzed with a FACSCalibur flow cytometer (BD). Data were processed with FlowJo software (Tree Star, Inc.).
qRT-PCR
Total RNA was extracted from 5 × 107 bloodstream form trypanosomes using TRIZOL reagent (Invitrogen) according to the manufacturer's recommendations. Reverse transcription was conducted for 30 min at 50°C with the SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative PCR assays were performed in an iCycler IQ real-time PCR detection system (Bio-Rad Laboratories), with iQ SYBR green super mix (Bio-Rad Laboratories) according to the manufacturer's recommendations. Primer sets were designed from the bloodstream ES library (http://www.genedb.org/genedb/tbrucei427) to specifically amplify telomeric VSG genes VSG121 (MITat 1.6), VSGVO2 (MITat 1.9), VSGBn2 (MITat 1.13), VSG224 (MITat 1.3), VSGJS1 (MITat 1.17), and VSG221 (MITat 1.2). See Table S1 for primer sequences ordered according to bloodstream ES nomenclature (Hertz-Fowler et al., 2008). Two VSG genes known to be BCs (VSG118-BC and VSG222-BC) were also amplified (Liu et al., 1983; Ansorge et al., 1999). The PI3K-like gene was used as a single copy gene control. Relative levels of gene expression were measured using the comparative cycle threshold CT method between samples and myosin 1B as a reference gene (iCycler IQ user bulletin; Bio-Rad Laboratories).
Online supplemental material
Fig. S1 shows the general characterization of sister chromatid separation in T. brucei. Fig. S2 shows the determination of T. brucei nuclear mitotic phases using DAPI staining. Fig. S3 shows that the depletion of essential proteins, including TbActin, TbMyosin 1B, and TbRPB5, does not result in changes in VSG switching frequency. Fig. S4 shows that knockdown of TbSCC1 results in an increase in the in situ switching-off rate of the VSG221 ES and switching-on rate of VSG121 ES by FACS analysis. Fig. S5 shows that knockdown of TbSCC1 promotes the appearance of VSGVO2 variants. Table S1 lists all of the primers used in the qRT-PCR analysis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200902119/DC1.
Acknowledgments
We thank M. Carrington (University of Cambridge, Cambridge, England, UK) for providing anti-TbSCC1 antiserum. We thank I. Vidal for excellent technical assistance, including generation of anti-VSG121 monoclonal antibodies. We also thank K. Gull (University of Oxford, Oxford, England, UK) for anti-BB antibody, G.A.M. Cross (The Rockefeller University, New York, NY) for anti-VSG221, and G. Rudenko (University of Oxford) for anti-VSGVO2.
This work was funded by Howard Hughes Medical Institute (HHMI) grant 55005525 and by Spanish Ministry of Science grant SAF2006-01763. D. Van Tyne is supported by a Fulbright grant from the Spanish Commission of Cultural, Educational, and Scientific Exchange. M. Navarro is an HHMI International Research Scholar.
Footnotes
Abbreviations used in this paper: BB, basal body; BC, basic copy; ES, expression site; ESB, ES body; IF, immunofluorescence; LUT, look up table; MITat, Molteno Institute Trypanozoon antigenic type; qRT-PCR, quantitative RT-PCR; rDNA, ribosomal DNA; VSG, variant surface glycoprotein.
References
- Aitcheson N., Talbot S., Shapiro J., Hughes K., Adkin C., Butt T., Sheader K., Rudenko G. 2005. VSG switching in Trypanosoma brucei: antigenic variation analysed using RNAi in the absence of immune selection.Mol. Microbiol. 57:1608–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge I., Steverding D., Melville S., Hartmann C., Clayton C. 1999. Transcription of ‘inactive’ expression sites in African trypanosomes leads to expression of multiple transferrin receptor RNAs in bloodstream forms.Mol. Biochem. Parasitol. 101:81–94 [DOI] [PubMed] [Google Scholar]
- Barquilla A., Crespo J.L., Navarro M. 2008. Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation.Proc. Natl. Acad. Sci. USA. 105:14579–14584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont A.S. 2001. Visualizing chromosome dynamics with GFP.Trends Cell Biol. 11:250–257 [DOI] [PubMed] [Google Scholar]
- Bessat M., Ersfeld K. 2009. Functional characterization of cohesin SMC3 and separase and their roles in the segregation of large and minichromosomes in Trypanosoma brucei.Mol. Microbiol. 71:1371–1385 [DOI] [PubMed] [Google Scholar]
- Borst P. 2002. Antigenic variation and allelic exclusion.Cell. 109:5–8 [DOI] [PubMed] [Google Scholar]
- Borst P., Ulbert S. 2001. Control of VSG gene expression sites.Mol. Biochem. Parasitol. 114:17–27 [DOI] [PubMed] [Google Scholar]
- Bulow R., Nonnengasser C., Overath P. 1989. Release of the variant surface glycoprotein during differentiation of bloodstream to procyclic forms of Trypanosoma brucei.Mol. Biochem. Parasitol. 32:85–92 [DOI] [PubMed] [Google Scholar]
- Chaves I., Zomerdijk J., Dirks-Mulder A., Dirks R.W., Raap A.K., Borst P. 1998. Subnuclear localization of the active variant surface glycoprotein gene expression site in Trypanosoma brucei.Proc. Natl. Acad. Sci. USA. 95:12328–12333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross G.A. 1984. Release and purification of Trypanosoma brucei variant surface glycoprotein.J. Cell. Biochem. 24:79–90 [DOI] [PubMed] [Google Scholar]
- Devaux S., Kelly S., Lecordier L., Wickstead B., Perez-Morga D., Pays E., Vanhamme L., Gull K. 2007. Diversification of function by different isoforms of conventionally shared RNA polymerase subunits.Mol. Biol. Cell. 18:1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. 2007. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes.Chromosoma. 116:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstler M., Boshart M. 2004. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei.Genes Dev. 18:2798–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersfeld K., Gull K. 1997. Partitioning of large and minichromosomes in Trypanosoma brucei.Science. 276:611–614 [DOI] [PubMed] [Google Scholar]
- Figueiredo L.M., Janzen C.J., Cross G.A. 2008. A histone methyltransferase modulates antigenic variation in African trypanosomes.PLoS Biol. 6:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss S.H., Omura M., Mombaerts P. 2007. Local and cis effects of the H element on expression of odorant receptor genes in mouse.Cell. 130:373–384 [DOI] [PubMed] [Google Scholar]
- Gluenz E., Sharma R., Carrington M., Gull K. 2008. Functional characterization of cohesin subunit SCC1 in Trypanosoma brucei and dissection of mutant phenotypes in two life cycle stages.Mol. Microbiol. 69:666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondor A., Ohlsson R. 2008. Chromatin insulators and cohesins.EMBO Rep. 9:327–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V. 2007. Sister chromatid cohesion: the cohesin cleavage model does not ring true.Genes Cells. 12:693–708 [DOI] [PubMed] [Google Scholar]
- Gunzl A., Bruderer T., Laufer G., Schimanski B., Tu L.C., Chung H.M., Lee P.T., Lee M.G. 2003. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei.Eukaryot. Cell. 2:542–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi M.A., Bochar D.A., Schmiesing J.A., Dong Y., Barak O.G., Speicher D.W., Yokomori K., Shiekhattar R. 2002. A chromatin remodelling complex that loads cohesin onto human chromosomes.Nature. 418:994–998 [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C., Figueiredo L.M., Quail M.A., Becker M., Jackson A., Bason N., Brooks K., Churcher C., Fahkro S., Goodhead I., et al. 2008. Telomeric expression sites are highly conserved in Trypanosoma brucei.PLoS ONE. 3:e3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield J.A., Anagnostou S.H., Hu J.K., Cho K.H., Geisler R., Lieschke G., Crosier K.E., Crosier P.S. 2007. Cohesin-dependent regulation of Runx genes.Development. 134:2639–2649 [DOI] [PubMed] [Google Scholar]
- Hughes K., Wand M., Foulston L., Young R., Harley K., Terry S., Ersfeld K., Rudenko G. 2007. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes.EMBO J. 26:2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Wang C.C. 2005. Depletion of anaphase-promoting complex or cyclosome (APC/C) subunit homolog APC1 or CDC27 of Trypanosoma brucei arrests the procyclic form in metaphase but the bloodstream form in anaphase.J. Biol. Chem. 280:31783–31791 [DOI] [PubMed] [Google Scholar]
- Landeira D., Navarro M. 2007. Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei.J. Cell Biol. 176:133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.Y., Van der Ploeg L.H., Rijsewijk F.A., Borst P. 1983. The transposition unit of variant surface glycoprotein gene 118 of Trypanosoma brucei. Presence of repeated elements at its border and absence of promoter-associated sequences.J. Mol. Biol. 167:57–75 [DOI] [PubMed] [Google Scholar]
- Lomvardas S., Barnea G., Pisapia D.J., Mendelsohn M., Kirkland J., Axel R. 2006. Interchromosomal interactions and olfactory receptor choice.Cell. 126:403–413 [DOI] [PubMed] [Google Scholar]
- Lopez-Rubio J.J., Gontijo A.M., Nunes M.C., Issar N., Hernandez Rivas R., Scherf A. 2007. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites.Mol. Microbiol. 66:1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A. 2007. Cohesin regulation: fashionable ways to wear a ring.Chromosoma. 116:321–329 [DOI] [PubMed] [Google Scholar]
- McKee B.D. 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis.Biochim. Biophys. Acta. 1677:165–180 [DOI] [PubMed] [Google Scholar]
- Nasmyth K. 2002. Segregating sister genomes: the molecular biology of chromosome separation.Science. 297:559–565 [DOI] [PubMed] [Google Scholar]
- Navarro M., Cross G.A. 1996. DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei.Mol. Cell. Biol. 16:3615–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M., Gull K. 2001. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei.Nature. 414:759–763 [DOI] [PubMed] [Google Scholar]
- Navarro M., Penate X., Landeira D. 2007. Nuclear architecture underlying gene expression in Trypanosoma brucei.Trends Microbiol. 15:263–270 [DOI] [PubMed] [Google Scholar]
- Ngo H., Tschudi C., Gull K., Ullu E. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei.Proc. Natl. Acad. Sci. USA. 95:14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbadoyi E., Ersfeld K., Robinson D., Sherwin T., Gull K. 2000. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis.Chromosoma. 108:501–513 [DOI] [PubMed] [Google Scholar]
- Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms.Cell. 132:422–433 [DOI] [PubMed] [Google Scholar]
- Pays E., Vanhamme L., Perez-Morga D. 2004. Antigenic variation in Trypanosoma brucei: facts, challenges and mysteries.Curr. Opin. Microbiol. 7:369–374 [DOI] [PubMed] [Google Scholar]
- Penate X., Lopez-Farfan D., Landeira D., Wentland A., Vidal I., Navarro M. 2009. RNA pol II subunit RPB7 is required for RNA pol I-mediated transcription in Trypanosoma brucei.EMBO Rep. 10:252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D., van Steensel B. 2008. Linking cohesin to gene regulation.Cell. 132:925–928 [DOI] [PubMed] [Google Scholar]
- Ralph S.A., Scherf A. 2005. The epigenetic control of antigenic variation in Plasmodium falciparum.Curr. Opin. Microbiol. 8:434–440 [DOI] [PubMed] [Google Scholar]
- Rollins R.A., Korom M., Aulner N., Martens A., Dorsett D. 2004. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene.Mol. Cell. Biol. 24:3100–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., Peacock L., Gluenz E., Gull K., Gibson W., Carrington M. 2008. Asymmetric cell division as a route to reduction in cell length and change in cell morphology in trypanosomes.Protist. 159:137–151 [DOI] [PubMed] [Google Scholar]
- Sheader K., Berberof M., Isobe T., Borst P., Rudenko G. 2003. Delineation of the regulated Variant Surface Glycoprotein gene expression site domain of Trypanosoma brucei.Mol. Biochem. Parasitol. 128:147–156 [DOI] [PubMed] [Google Scholar]
- Siegel T.N., Hekstra D.R., Cross G.A. 2008. Analysis of the Trypanosoma brucei cell cycle by quantitative DAPI imaging.Mol. Biochem. Parasitol. 160:171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. 2001a. Chromosome cohesion and segregation in mitosis and meiosis.Curr. Opin. Cell Biol. 13:754–761 [DOI] [PubMed] [Google Scholar]
- Uhlmann F. 2001b. Secured cutting: controlling separase at the metaphase to anaphase transition.EMBO Rep. 2:487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Figueiredo L.M., Espinal A., Okubo E., Li B. 2009. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei.Cell. 137:99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor.Nature. 451:796–801 [DOI] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C., Cross G.A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei.Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- Woodward R., Gull K. 1990. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei.J. Cell Sci. 95:49–57 [DOI] [PubMed] [Google Scholar]