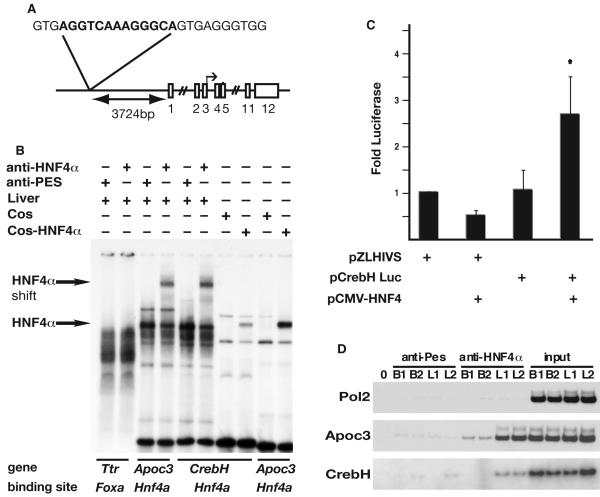
Fig. 3. CrebH is a direct target of HNF4α transcriptional activity.
(A) Schematic showing the genomic location and sequence of the identified HNF4α binding site relative to CrebH (exons shown as boxes). (B) The ability of HNF4α protein to bind the putative HNF4α-binding site was confirmed by EMSA. Radiolabeled oligonucleotides representing binding sites were incubated with liver nuclear extracts in the presence of anti-HNF4α antibody, which resulted in a retarded migration of HNF4α-bound complexes (arrows), or anti-Pes1 antibody (negative control). Alternatively, EMSAs were performed using nuclear extracts from COS-7 cells or COS-7 cells expressing HNF4α. A previously described HNF4α binding site in the Apolipoprotein c3 (Apoc3) promoter served as a positive control and a Foxa (Hnf3) binding site within the Transthyretin (Ttr) promoter served as a negative control. (C) 293T cells were transfected with plasmids in which expression of luciferase was driven by the HIV basal promoter (pZLHIVS) or in addition a 207bp fragment from the CrebH gene that contained the HNF4α binding site (pCrebH-Luc1) in the presence or absence of exogenously expressed HNF4α. Luciferase levels from five independent experiments are presented as fold difference relative to cells transfected with pZLHIVS alone. Significance was determined by Student’s t-test (p<0.05). D) ChIP analyses were performed on chromatin extracted from two independent E18.5 livers (L1, L2) or brains (B1, B2), which acted as a negative control tissue that does not express HNF4α. Chromatin was precipitated using anti-HNF4α or anti-Pes1 (negative control), and specific primers were used to amplify input chromatin or chromatin precipitated from the Pol2 promoter (negative control), Apoc3 promoter (positive control), or CrebH.