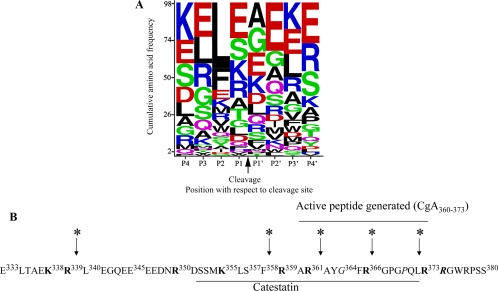
Figure 6.
CTSL cleavage sites in CgA. A, Bioinformatic analysis. Recombinant CgA (10 μm) was digested with CTSL (0.3 μm) for 15 min and subjected to LC-MS/MS analyses. The 98 resulting cleavage sites (P1 and P1′ positions) and the flanking sequences (supplemental Tables 1 and 2) were combined into a 20 × 8 amino acid frequency matrix. WebLogo was used to generate a cumulative frequency sequence from this matrix. Amino acids are arranged from top to bottom in order of descending frequency, with the height of each letter proportional to its frequency. B, Depiction of preferred cleavage sites of CTSL within the catestatin region of human CgA. Catestatin region within human CgA was shown as underlined CgA352–372. All the monobasic and paired dibasic sites were shown in bold. The predicted cleavage sites were shown by arrow. The catestatin variant positions were shown in italics.