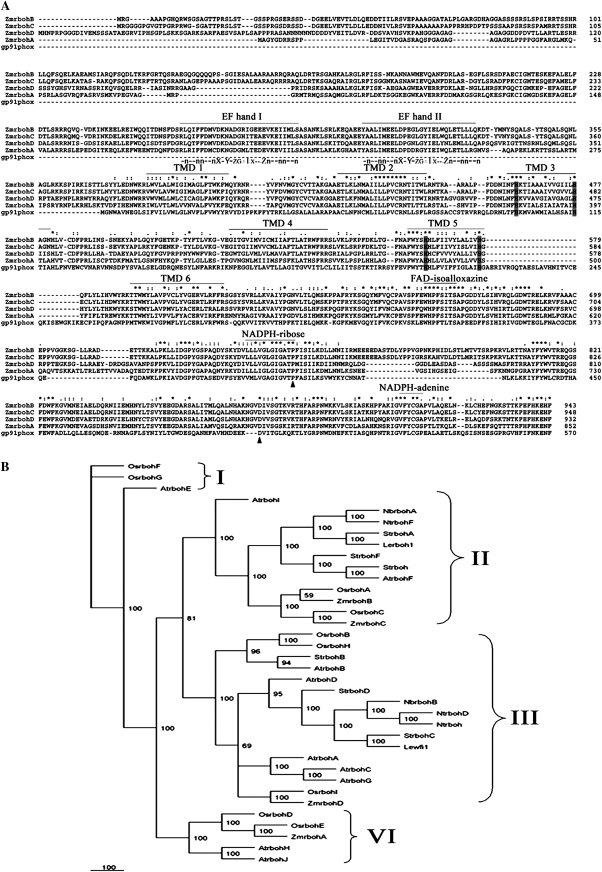
Fig. 1.
Alignment and phylogenetic relationship of ZmrbohA–D and its related members of the rboh family. (A) Alignment of the predicted amino acid sequences of ZmrbohA–D and human gp91phox. Multiple alignment of the predicted ZmrbohA–D and gp91phox protein was made with the CLUSTAL-W program. Asterisks (*) indicate the strictly conserved amino acid residues. Amino acid similarity (: and .) is based on the CLUSTAL-X convention and dashes indicate gaps in the sequence to allow for maximal alignment. Six potential transmembrane-spanning domains (TMD 1 to TMD 6) are indicated with overlines. Histidine residues involved in haem binding are boxed in grey scale. Solid triangles under the sequences indicate amino acid residues that are required for the human NADPH oxidase function and conserved between gp91phox and the rboh proteins. EF hand motifs in the N-terminal domain of rboh are overlined. Beneath these motifs is the sequence of a canonical EF hand. n is usually a hydrophobic residue. Dashes indicate variable amino acid residues. X, Y, Z, and -X, contain oxygen within their side chains. Carbonyl oxygen of # serves as a ligand. -Z is usually glutamic acid. (B) Unrooted phylogenetic tree of various plant rbohs. Bootstrap values of 50% or higher are shown on significant nodes. Species names: At, Arabidopsis thaliana; Le, Lycopersicon esculentum; Nb, Nicotiana benthamiana; Nt, Nicotiana tabacum; Os, Oryza sativa; St, Solanum tuberosum; Zm, Zea mays.