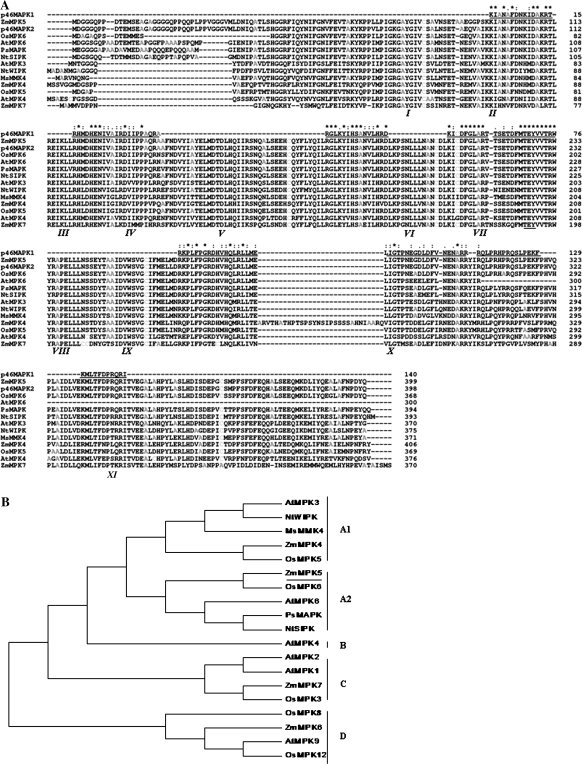
Fig. 9.
Comparison of the maize p46MAPK with other MAPKs from higher plants. (A) Alignment of deduced amino acid sequence of the maize p46MAPK (p46MAPK2) with those of other members of the MAPK family. Asterisks (*) shows the strictly conserved amino acid residues. Amino acid similarity (·) is based on the CLUSTAL-X convention and dashes indicate gaps introduced to maximum alignment. The conserved TEY phosphorylation motif for MAPK is marked with the dotted line under the sequences. Roman numerals indicate the 11 major conserved subdomains of serine/threonine protein kinases. The peptide sequences obtained by microsequencing of the purified protein (called p46MAPK1) are shown above the ZmMPK5 sequence. (B) Phylogenetic relationship of ZmMPK5 with other plant MAPKs. Phylogenetic tree based on the genetic distance of the protein sequences was constructed by the neighbor-joining method using MEGA 3.1 software. Species names: Zm, Zea mays; At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Os, Oryza sativa; Ps, Pisum sativum; Ms, Medicago sativa.