Abstract
In this work we demonstrate how polarized light absorption spectroscopy (linear dichroism (LD)) analysis of the peptide ultraviolet-visible spectrum of a membrane-associated protein (cytochrome (cyt) c) allows orientation and structure to be assessed with quite high accuracy in a native membrane environment that can be systematically varied with respect to lipid composition. Cyt c binds strongly to negatively charged lipid bilayers with a distinct orientation in which its α-helical segments are on average parallel to the membrane surface. Further information is provided by the LD of the π-π∗ transitions of the heme porphyrin and transitions of aromatic residues, mainly a single tryptophan. A good correlation with NMR data was found, and combining NMR structural data with LD angular data allowed the whole protein to be docked to the lipid membrane. When the redox state of cyt c was changed, distinct variations in the LD spectrum of the heme Soret band were seen corresponding to changes in electronic transition energies; however, no significant change in the overall protein orientation or structure was observed. Cyt c is known to interact in a specific manner with the doubly negatively charged lipid cardiolipin, and incorporation of this lipid into the membrane at physiologically relevant levels was indeed found to affect the protein orientation and its α-helical content. The detail in which cyt c binding is described in this study shows the potential of LD spectroscopy using shear-deformed lipid vesicles as a new methodology for exploring membrane protein structure and orientation.
Introduction
Biological lipid membranes harbor a significant portion of the total number of proteins encoded for by the genome. These proteins often have vital functions, such as maintaining cell homeostasis, cell-cell signaling, and cellular response to external stimuli (1). Membrane proteins (in the form of receptors) are also the most common cellular drug targets for pharmaceuticals on the market today (2). Thus, research on membrane proteins is important not only in the context of understanding cellular function, but also in the hunt for new or improved drug leads. However, solving membrane protein structures has proven difficult. Although NMR spectroscopy and x-ray crystallography have been used successfully to obtain atomic-resolution structure data for water-soluble proteins, it is evident from the records in the Protein Data Bank (PDB) (3) that the number of membrane protein structures is lagging far behind. As of December 2007, the structures of 178 unique membrane proteins and peptides had been deposited in the Membrane PDB, as compared with a total number of ∼50,000 protein structures deposited in the PDB (4). There are several reasons for this discrepancy, including experimental difficulties associated with expressing membrane proteins and subsequently reconstituting them in membrane-mimicking environments in their functional state. Also, obtaining diffracting crystals of membrane proteins has proven very difficult and sometimes requires rather harsh treatment of the sample, including excessive addition of detergents. In addition, the membrane-mimicking environments used for protein crystallization are often substantially different from a lipid bilayer, and consequently there is a certain risk that the solved structure will not represent the functional state of the protein. Finally, and most importantly in our context, since membrane proteins are crystallized without their natural lipid bilayer, all orientational and membrane associative information is missing. In principle, this information may be available from solution NMR experiments on proteins embedded in membrane fragments (e.g., “bicelles”), provided that the total system is small enough to display a short orientation correlation time on the NMR timescale (5).
In this study we explore the use of polarized light absorption spectroscopy (linear dichroism (LD)) to study the orientation and binding geometry of a membrane-associated protein hosted in the bilayer of a large (100 nm in diameter) unilamellar lipid vesicle. The methodology for studying the orientation of membrane-associated molecules with the use of shear-deformed lipid vesicles was developed in our laboratory 10 years ago (6), and since then the technique has been improved with respect to sensitivity and applications (7–13). Here we demonstrate, using cytochrome (cyt) c as a model, that it is possible to obtain detailed orientational information about a membrane-associated protein from LD of specific electronic transitions within the protein. We also show the usefulness of this technique by confirming that the orientational information obtained from LD can be exploited to dock an NMR solution structure of cyt c onto a lipid membrane, thereby creating an “atomic-resolution model” that is in agreement with earlier speculated orientations of the protein. Finally, we prove that the LD technique is sensitive enough to probe small structural changes within the protein induced, in this case, by external stimuli.
Cyt c is a small globular redox protein (104 residues, ∼34 Å in diameter) with an α-helical content of ∼30–40% (14,15). The protein is normally found in the intermembrane space of mitochondria, where it participates in the mitochondrial respiratory chain by transferring electrons between the membrane protein complexes CoQH2-cyt c reductase and the cyt c oxidase complexes (1). Cyt c is a readily water-soluble protein, but it binds strongly to acidic lipid membranes, particularly at low ionic strength conditions (14,16), and it has traditionally been considered as diffusing in two dimensions at the surface of the mitochondrial inner membrane between its redox partners (17). In addition, cyt c translocates across the mitochondrial membrane to be released into the cytosol as a crucial step in the early stages of apoptosis (18). Mitochondrial membranes are rich in negatively charged lipids, and particularly in the biphosphatidyl glycerol lipid cardiolipin (CL), which is a dimeric phospholipid with two phosphatidyl moieties linked by a central glycerol group. The electrostatic interaction between cyt c and negatively charged lipids has been suggested to occur at a lipid-binding site on the protein surface (19,20). In addition, a specific interaction between cyt c and CL that results in alterations in both protein secondary structure and degree of penetration into the bilayer has been suggested (21).
Cyt c is ideal as model protein for extending the flow LD technique from studies of model peptides to full-length proteins because it is of a manageable size, its structure and behavior have been extremely well studied (both in solution and in the presence of lipid membranes), and its water-soluble state has been characterized at atomic resolution (22,23). Moreover, it contains several useful chromophores with characteristic absorption in the ultraviolet (UV)-visible region of the spectrum, including the heme porphyrin group, a single tryptophan residue (Trp59), and four tyrosines. Also, the redox state of cyt c can be changed simply by the addition of a reducing agent, and we saw a possibility to explore structural changes associated with the electron transfer process. Previous studies of cyt c indicated differences between the two redox states, including heme distortions and different orientations of the axial histidine (22,24).
This study shows that the binding geometry of membrane-associated cyt c can be qualitatively determined with the use of LD spectroscopy, a technique that is relatively simple in terms of experimental procedures and can be straightforwardly adapted to any commercially available CD spectropolarimeter. High-quality orientational information was obtained for certain protein motifs, and the obtained LD data were used to build a model of the membrane-bound protein from an existing atomic-resolution structure of the soluble protein, verifying a near-identical fold and the existence of one membrane-bound state.
Materials and Methods
Chemicals
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG) were purchased from Larodan (Malmö, Sweden). Sucrose (99.5% pure), horse heart cyt c (100% pure), CL (98% pure, from 67 bovine hearts), and sodium dithionite (reducing agent) were purchased from Sigma-Aldrich (St. Louis, MO). The buffer contained 10 mM phosphate (pH 7.0) and was prepared in ultrapure water. In all LD experiments, the buffer contained 50% (w/w) sucrose (see below).
Preparation of lipid vesicles
A chloroform solution with the desired lipid composition was evaporated to dryness in a round-bottomed flask using a rotary evaporator. The resulting lipid film was then put under reduced pressure over night to ensure complete removal of chloroform. The lipid film was dispersed in buffer, and monodisperse large unilamellar vesicles (hereafter denoted lipid vesicles) were obtained by freeze-thawing (five times) followed by subsequent extrusion (21 times) through Nucleopore polycarbonate filters (100 nm diameter) using an extruder (Lipofast-Pneumatic, Avestin, Ottawa, Ontario, Canada).
Binding assay
The extent of oxidized cyt c binding to the lipid membrane was measured with the use of ultracentrifugation to separate free and membrane-bound protein (25). Briefly, lipid vesicles (5 mM total lipid concentration) were incubated with cyt c at a protein/lipid ratio of 1:200 and equilibrated for 3 h at room temperature. The lipid vesicle-protein complexes were separated from free protein by ultracentrifugation for 4 h at 4°C (50,000 rpm, TLA120:2 rotor; Beckman Optima Max Ultracentrifuge). The free protein concentration was determined spectrophotometrically by measuring the absorbance (at 410 nm) in the supernatant. The extinction coefficient used for oxidized cyt c was ɛ410 = 101600 M−1cm−1 (14).
Circular dichroism
Circular dichroism (CD) was used to examine the secondary structure of free and bound cyt c. Spectra were recorded on a Jasco J-810 spectropolarimeter thermostated at 25°C in 1 nm increments, between 190 and 250 nm to explore protein secondary structure, and between 325 and 510 nm to detect CD from the heme group. The scan speed was 50 nm/min, the response was 0.5 s, and the band pass was 1 nm. Ten to 20 scans were accumulated and averaged by the computer. All spectra were corrected for background contributions by subtracting appropriate blanks. The maximum lipid concentration was 1 mM to avoid spectral distortions due to excessive light scattering. The protein/lipid ratio was 1:200, the same as that used in the LD experiments (see below). Secondary structure analysis was performed using the CDSSTR algorithm (26–28) accessed through the online server DICHROWEB (29,30).
Linear dichroism
LD is defined as the difference in absorption of linearly polarized light oriented parallel and perpendicular to a macroscopic orientation axis:
| (1) |
Lipid vesicles can be deformed into an ellipsoidal shape and aligned by shear flow in a Couette cell device, rendering absorbing transition moments in membrane-associated protein molecules oriented and thus amenable to exhibit LD (6). Chromophores with absorbing transitions whose moments are parallel to the lipid surface exhibit positive LD, and transitions with moments oriented along the bilayer normal exhibit negative LD. The orientation of a specific transition moment can be assessed from its reduced LD, LDr. This quantity is obtained by normalizing the LD with respect to the isotropic absorption, Aiso, which should be recorded on the same sample, preferably in a cell with the same pathlength as the Couette cell. The LDr is a concentration-independent variable, and for a pure nonoverlapping absorption band corresponding to transition moment i, the LDr is related to the latter's angle relative to the membrane normal, αi, according to:
| (2) |
where S is the macroscopic orientation parameter that describes the overall alignment of the sample. A perfectly oriented sample exhibits S = 1, whereas for an isotropic (random) system S = 0. S is principally dependent on two factors: 1), the deformation and thus the degree of alignment of the lipid vesicles; and 2), the microscopic ordering of the lipids (acyl chains) within the vesicle membrane (6,7).
Samples for LD were prepared by addition of a freshly made aqueous cyt c solution to a final concentration of 25 μM to a 5 mM lipid vesicle dispersion (giving a protein/lipid ratio of 1:200), followed by 3 h incubation at ambient temperature. The buffer used contained 50% (w/w) sucrose to reduce light scattering from the lipid vesicles (8). This high-viscosity buffer also increases the viscous drag in the Couette cell, which results in a higher degree of deformation of the lipid vesicles and hence improved overall orientation of the samples. LD spectra were recorded on a Jasco J-720 spectropolarimeter equipped with an Oxley prism to obtain linearly polarized light (31), using an in-house-built outer-rotating quartz Couette flow cell with a total pathlength of 1 mm. The samples were aligned at a shear flow of 3100 s−1. Spectra were recorded at room temperature between 200 and 700 nm with a band pass of 2 nm and a scan speed of 100 nm/min. Three spectra were accumulated and averaged by the computer. All spectra were corrected for background contributions by subtracting a baseline spectrum recorded on the same sample but without rotation of the Couette cell.
Isotropic absorbance spectra (Aiso) were collected on all LD samples using a Varian Cary4 spectrophotometer and a 1 mm quartz cell. The macroscopic orientation parameter, S, was determined for each sample using retinoic acid as the membrane probe as previously described (11,13). Retinoic acid was added to LD samples after the protein LD spectrum had been recorded to a final concentration of 25 μM from a 10 mM stock solution. Samples were incubated for an additional 45 min before measurement to allow the retinoic acid to bind to the lipid vesicles in its intercalated state (11). Retinoic acid binds to lipid membranes with a high degree of ordering and aligns along the lipid chains, with the carboxylic acid group remaining at the membrane interface. It is a good membrane probe with peptides or proteins experiments because its main absorption band is centered around 350 nm and thus does not overlap significantly with the absorption of protein aromatic side chains or the absorption in the heme Soret band (11,32). S was calculated for each individual sample from the LDr of retinoic acid under the assumption that the retinoic acid transition moment is oriented perfectly perpendicular to the lipid surface (the lipid vesicle orientation axis) and hence parallel to the membrane normal, thus setting αretinoic acid to 0° in Eq. 2. This assumption is reasonable because retinoic acid exhibits the highest LDr value that we have managed to record to date using flow-oriented lipid vesicles (11). We previously reported that S varies considerably with lipid composition (12), and we and others have noticed that S seems to vary slightly between lipid vesicles that have identical compositions but were prepared from different lipid films (11,32). The latter effect seems inherent to the preparation method and is not affected or varied by the addition of protein; however, as a result, protein LD spectra recorded under identical conditions but in different lipid vesicle batches can have different magnitudes. We found that this discrepancy can be satisfactorily corrected for by normalizing each LD spectrum with respect to S of that particular sample. This has been done in Figs. 2–5
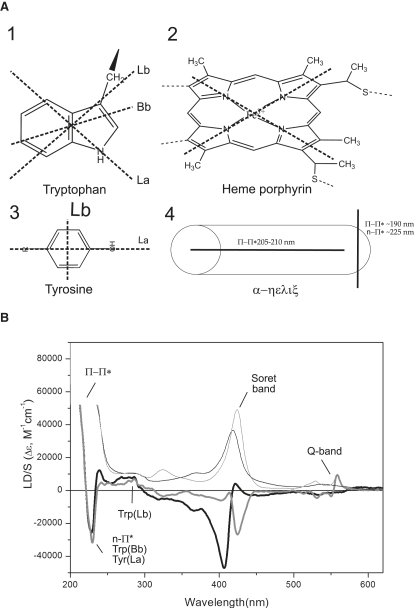
Figure 2.
(A) Four main intrinsic chromophores in cyt c: 1), the indole chromophore of tryptophan with transition moments Lb at ∼290 nm, La at ∼270 nm, and Bb at ∼225–230 nm (37); 2), the heme porphyrin prosthetic group with the x and y polarizations of the Soret band (33); 3), the tyrosine with its two perpendicular transition moments, the Lb at 276 nm, and the La at ∼230 nm (59); and 4), the peptide α-helix (40,41). (B) LD spectra of oxidized (black bold line) and reduced (gray bold line) cyt c associated to POPC/POPG (60:40) lipid vesicles. The corresponding absorption spectra (divided by a factor of 3) are included for comparison (oxidized as black thin line and reduced as gray thin line).
Figure 3.
Schematic of the TEM for stepwise reduction of the Soret transitions. The coefficient α (Eq. 3) is varied and the combinations that most accurately recreate the spectral features of the Soret bands are shown in bold with indicated Ldr values.
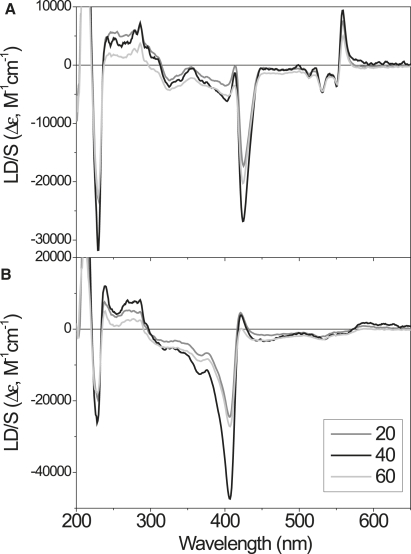
Figure 4.
LD spectra of oxidized (A) and reduced (B) cyt c in POPC/POPG lipid vesicles containing 20, 40, and 60 mol % negatively charged POPG.
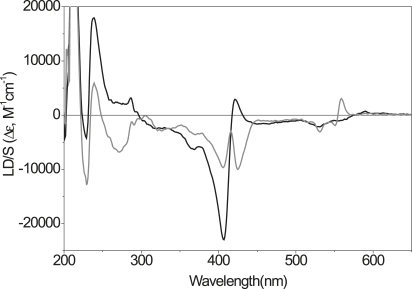
Figure 5.
LD spectra of oxidized (black line) and reduced (gray line) cyt c bound to CL-containing lipid vesicles (POPC/CL (80:20)).
The CD spectropolarimeter records spectra in units of ellipticity (θ, mdeg). This unit has no meaning in terms of LD, but can be easily converted into differential absorption expressed in molar absorptivity units according to:
| (3) |
where c is the protein concentration in the sample, and l is the pathlength of the Couette cell. All spectra presented in Figs. 2–4 were first converted to Δɛ and then normalized with respect to S, and are therefore expressed in units of M−1cm−1.
Transition moments in cyt c
The structures of the major chromophores in cyt c (the heme group, the tryptophan side chain, the tyrosine side chain, and the peptide α-helix) are given in Fig. 2 A. The arrows in the figure show the directions of the main transition moments in each chromophore, and their approximate absorption maxima are indicated in the figure legend and outlined below.
The aromatic tryptophan side-chain chromophore, indole, has three strongly absorbing transitions: the La and Lb transitions, which overlap in the 250–300 nm region, and the Bb transition at 225–230 nm (hereafter denoted La(trp), Lb(trp), and Bb(trp), respectively). La displays a broad and unstructured absorption profile centered on 270 nm, and Lb displays structured absorption characterized by two distinct vibronic peaks just below 290 nm. The tyrosine side chain has two main absorbing transitions whose moments are polarized perpendicular to each other: Lb with an absorption maximum at 276 nm, and La, which absorbs at ∼230 nm (hereafter denoted Lb(tyr) and La(tyr)).
The heme group has two nearly degenerate π→π∗ transitions whose moments are polarized perpendicular to each other in the heme x,y plane (33). The actual direction of the two transitions in the plane of the heme are not known but have been proposed to lie in the x- and y-Fe-pyrrole ring direction (34,35). They absorb at ∼430 nm (the Soret band). The heme group displays absorption also at ∼550 nm (the Q-band), but only in the reduced state. The orientation of the transition moments contributing to the absorption in the Q-band are not known.
The peptide bond absorbs light in the far-UV region of the spectrum, and the relevant transitions for an LD experiment are the π→π∗ transitions centered around 200 nm and the forbidden and therefore only weakly absorbing n→π∗ transitions around 220–230 nm. In an α-helix the π→π∗ transitions interact through exciton coupling and give rise to a resultant transition polarized parallel to the helix axis that absorbs at 210 nm, and a resultant transition polarized perpendicular to the helix axis that absorbs at 190 nm. It is not possible to resolve the latter with our experimental setup. The n→π∗ transitions are oriented on average perpendicular to the helix axis.
Estimation of LDr values and insertion angles
The relative contribution of overlapping transition moments to the LD spectrum in the “aromatic” (250–300 nm) region where the tryptophan and tyrosine side chains absorbs, and in the Soret band (∼430 nm) where the heme group absorbs were obtained using the trial and error method (TEM) of stepwise spectral reduction originally developed by Thulstrup et al. (36). TEM relies on eliminating the contribution of a specific spectral feature from the LD spectrum and makes use of the fact that the overlapping transitions have different spectral profiles. The performance of TEM depends on how easily the overlapping spectral profiles can be distinguished. The alternative way to extract LDr values from LD spectra is to reconstruct the LD signature in the wavelength region of interest with the help of the spectral profiles of each absorption band that contributes to the LD spectrum using a least-squares fitting approach. However, TEM is significantly less sensitive to spectral noise because it does not aim to fit each point in the spectrum, and was therefore chosen to derive the LDr.
TEM can be described as follows: If x and y are the two absorption profiles that build up the absorption, A = x + y, LD is related to the absorption accordingly, LD = αx + βy, where α and β correspond to the LDr. Using trial values of α on the left-hand side of Eq. 4 below, the value is chosen that most closely eliminates the spectral contribution from transition x (and vice versa for Eq. 5):
| (4) |
| (5) |
The LDr of the Lb(trp) transition moment was derived from the LD and absorption spectra in the 250–310 nm region under the criterion that a successful TEM elimination of Lb(trp) was obtained when the two peaked vibronic signatures from the Lb transition at 290 nm disappeared in the spectral combination.
The LDr of the two Soret transitions was derived from the LD and absorption spectra in the 350–480 nm region. The orthogonally polarized x and y components that result from lifting the degeneracy of the Soret band were assumed to be of equal oscillator strength, and the components were resolved by Eqs. 3 and 4 by finding the smallest values of |α| and |β| that gave the x and y component equal areas. The resolvation procedure of the Soret transitions is illustrated in Fig. 3.
Construction of a cyt c binding model
The insertion angles of specific transition moments obtained from the LD experiments were used as constraints to dock two solution NMR structures of oxidized and reduced cyt c, respectively, onto a model lipid membrane, resulting in an “atomic resolution” model for the orientation of membrane-bound cyt c. The structural coordinates were obtained from the Research Collaboratory for Structural Bioinformatics PDB (accession codes 1AKK (22) and 1GIW (23) for the oxidized and reduced structures, respectively). Only those transition moments whose orientation could be unambiguously assigned were used to construct the model. These were the Lb(trp) transition moment and the two orthogonal x- and y-Soret transitions in the heme group. Since the directions of the latter transitions relative to the molecular framework of the heme group have not been experimentally assigned, we defined instead the LDr of the normal to the heme plane. This LDr value can be calculated from the LDr values of the x- and y-Soret transitions since the sum of the LDr values of three purely polarized orthogonal transitions in a chromophore must be zero (this follows from LDr(i) = 3Sii, for a transition polarized parallel with orientation axis i, i = x,y,z, and α Sii = 0, for a uniaxially oriented system). The binding model was constructed as follows: two vectors describing the directions of Lb(trp) and the normal of the heme plane were defined using appropriate coordinates in the .pdb file (the vector for the heme normal was obtained as the cross product of the x- and y-pyrrole-Fe-N directions of rings A and B; for directions of Lb(trp), refer to Albinsson and Norden (37)). The two vectors were simultaneously rotated first around the z axis and then around the x axis, using a minimization algorithm in MATLAB software (The MathWorks, Natick, MA) to find the orientation that best corresponds to the experimentally obtained orientation of Lb(trp) and the x- and y-Soret transition moments. The rotation angles that gave the best fit were then used to rotate all protein coordinates in the .pdb file, and models of membrane-bound cyt c, in its correct orientation, were drawn using the SwissPDBViewer and PyMol software.
Results
Cyt c binding affinity
The binding affinity of cyt c to lipid vesicles of relevant composition was determined using ultracentrifugation to separate bound and free protein. The fraction of unbound protein was estimated by absorbance measurements on the supernatant, and the bound fraction was derived under the assumption that the remaining protein fraction was membrane-bound. Table 1 shows values for the bound fraction, Xb, of oxidized cyt c in samples prepared in the same way as in the LD studies and at the same total concentrations (protein/lipid ratio of 1:200 and total lipid concentration of 5 mM). The degree of binding is high in all types of samples tested. The increase in Xb with increasing degree of negatively charged lipid points to that electrostatic attraction is clearly a major determinant for binding. Two physiologically relevant membrane levels of negatively charged CL lipid (38)—10 and 20 mol %—were also investigated. The binding affinity is somewhat lower in CL-containing vesicles compared to POPC/POPG vesicles prepared at comparable surface charge ratios (CL carries two negative charges, and 10 mol % CL is thus equivalent to 20 mol % POPG).
Table 1.
Fraction of bound cyt c and estimated α-helical content
| Bound fraction (Xb)∗ |
Secondary structure† |
||||
|---|---|---|---|---|---|
| Oxidized |
Oxidized |
Reduced |
|||
| Helix | Strand | Helix | Strand | ||
| POPC/POPG (80:20) | 0.82 | 0.39 | 0.10 | 0.39 | 0.25 |
| POPC/POPG (60:40) | 0.96 | 0.35 | 0.12 | 0.39 | 0.25 |
| POPC/POPG (40:60) | 0.95 | 0.35 | 0.12 | 0.40 | 0.27 |
| POPC/CL (90:10) | 0.70 | 0.18 | 0.35 | 0.16 | 0.40 |
| POPC/CL (80:20) | 0.88 | 0.20 | 0.26 | 0.33 | 0.16 |
| Free cyt c | — | 0.31 | 0.15 | 0.38 | 0.09 |
Fraction of bound protein was determined using an ultracentrifugation assay to separate free and bound protein.
Fraction of α-helix and β-strand secondary structure as determined by the CDSSTR algorithm applied to the recorded CD spectra in the 200–250 nm region, using reference set number 7 (includes 48 proteins) in DICHROWEB (29,30).
CD and secondary structure of cyt c
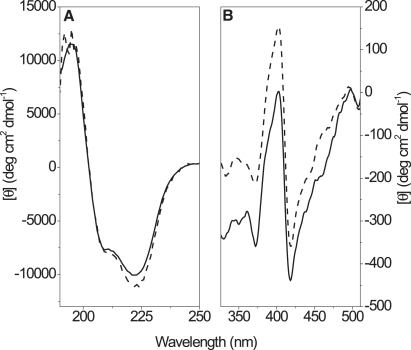
Fig. 1 shows CD spectra of oxidized cyt c in solution (10 mM phosphate buffer) and associated to POPC/POPG (60:40) lipid vesicles dispersed in the same buffer. The shape of the CD spectra confirms that cyt c is adopting a mainly α-helical conformation, which is in agreement with existing x-ray and NMR structures of cyt c in solution (22,23,39). The free and bound spectra are similar, but not identical, in both the UV/near-UV region where the spectrum is dominated by the peptide bond transitions, and the Soret band where the heme group absorbs. The CDSSTR algorithm was used to deconvolute the CD spectrum of each sample to estimate the secondary structure content. The analysis showed that the protein folds into mainly α-helical and “random coil” segments, but with some contribution of β-sheet structure. The estimated α-helical and strand contents are given in Table 1. The spectra in Fig. 1 represent the sample in which the largest deviation in secondary structure content between bound and free protein was observed (using the POPC/POPG lipid system). As can be seen from the data in Table 1, the variations in secondary structure content are minor, and therefore it is likely that cyt c largely retains its fold upon binding to POPC/POPG lipid vesicles. By contrast, when cyt c binds to lipid vesicles containing CL, the α-helical content is in general significantly lower. This indicates that cyt c may become partially unfolded upon interaction with membranes containing this particular lipid.
Figure 1.
CD spectra of oxidized cyt c free in solution (solid lines) and associated to POPC/POPG (40:60) lipid vesicles (dashed lines). The protein/lipid ratio was 1:200, and the lipid concentration was 1 mM. (A) The peptide bond spectral region. (B) The Soret spectral region of the heme group.
LD of membrane-bound cyt c—comparison of the oxidized and reduced forms
Fig. 2 B shows LD and isotropic absorbance spectra of oxidized and reduced cyt c bound to lipid vesicles containing 40 mol % negatively charged lipids (POPC/POPG (60:40)). The spectra are very different, particularly in the visible region (heme group absorption) but also in the aromatic region (tryptophan and tyrosine absorption). The LD in the Soret band (∼430 nm) is bisignate, reflecting the orthogonal orientations of the x- and y-heme transitions. In the oxidized spectra there is a large negative peak at shorter wavelengths and a small positive peak at longer wavelengths. Reduction of the heme group results in a sign shift of this split LD signal, with concurrent changes in magnitude of the two bands, and also appearance of LD and structured absorption in the Q-band (∼550 nm). Fig. 2 shows the LDr values of these transitions, which were estimated using TEM data analysis as described in Materials and Methods. An example of the TEM data analysis is shown in Fig. 3. Spectral differences are also observable in the 220–300 nm region, indicating some changes in orientation of aromatic side chains (see below). By contrast, the oxidized and reduced spectra are similar in the UV region and display strong positive peaks of nearly equal magnitudes around 208–210 nm (the peaks were truncated to better visualize other spectral features, but their maxima are at 230,000 M−1cm−1 and 225,000 M−1cm−1 for the oxidized and reduced form, respectively). These peaks emanate from the low-energy component of the peptide bond π→π∗ exciton coupling, which is observed for α-helical peptide segments according to Moffitt theory (40,41). These transition moments form a resultant transition oriented parallel to the long axis of α-helices (see Fig. 2 A and Materials and Methods). Therefore, the positive LD at 205–210 nm shows that the α-helical segments in cyt c are on average more parallel than perpendicular to the lipid bilayer. The magnitude of the signal indicates that the α-helices are well aligned along the membrane surface. The peaks in the oxidized and reduced spectra have, within experimental error, the same magnitude, suggesting that the overall orientation of the protein is not significantly altered by reduction of the heme group.
In addition to the π→π∗ transitions, each peptide bond amide chromophore has an n→π∗ transition, and in an α-helix these will give rise to a transition moment oriented perpendicular to the helix axis. This resultant transition moment absorbs at 220–225 nm but is only weakly allowed, and the extinction coefficient is thus very low (ɛ ∼ 100 M−1cm−1 per residue). However, the appearance of positive LD in the π→π∗ absorption band infers that the n→π∗ absorption band should be negative since their corresponding transitions are orthogonal (see Fig. 2 A). There is indeed a negative absorption band around 220–230 nm in both spectra in Fig. 2 B, but it cannot be assigned solely to the n→π∗ transition since the Bb(trp) and La(tyr) also absorb in this region.
Cyt c contains one single tryptophan residue (Trp59), which in principle makes it possible to assign its exact orientation with three angular coordinates (from the orientation of its Bb, La, and Lb transition moments). Both the oxidized and reduced spectra in Fig. 2 B display two small positive peaks just below 290 nm, which is a characteristic signature of the Lb(trp) absorption profile. Therefore, Trp59 must be oriented so that the Lb transition moment is more parallel than perpendicular to the membrane surface. The exact orientation of the Lb(trp) transition moment was obtained from TEM data analysis of the LD and absorption spectra in the aromatic region as detailed in Materials and Methods. The LDr values and calculated insertion angles are given in Table 2. The analysis shows that Lb(trp) is essentially perpendicular to the membrane normal (α = 87°) in both oxidized and reduced cyt c. The resultant LD in the 250–300 nm, obtained after subtraction of the Lb(trp) contribution, corresponds to the average orientation of the single La(trp) transition moment in tryptophan and the Lb(tyr) transition moments in the four tyrosine residues of cyt c. The resultant LD spectrum was not further resolved, but for the purpose of discussion of the change in LD in the aromatic region upon reduction of cyt c, a residual LDr value representing this average is included in Table 2. The orientation of La(trp) can be obtained once the binding geometry of the entire cyt c protein has been assigned. The relative contribution to LD in this region from the La(trp) and Lb(tyr) will be further considered in the Discussion.
Table 2.
Macroscopic orientation parameters (S), LDr values, and insertion angles (α) relative to the membrane normal for cyt c transition moments
| LDr/(S × Xb)† |
α |
||||||
|---|---|---|---|---|---|---|---|
| S∗ | Chromophore | Transition moment | Oxidized | Reduced | Oxidized | Reduced | |
| POPC/POPG (60:40) | 0.042 | Trp | Lb | +0.74 | +0.74 | 87° | 87° |
| La(trp)/Lb(tyr)‡ | +0.30 | 0 | 63° | 54° | |||
| heme | Soreti | −0.32 | +0.67 | 42° | 66° | ||
| Soretii | +0.74 | −0.42 | 87° | 46° | |||
| POPC/CL (80:20) | 0.050 | Trp | Lb | +0.57 | −0.79 | 74° | 34° |
| La(trp)/Lb(tyr) | +0.23 | −1.47 | 61° | 6° | |||
| Heme | Soreti | −0.29 | n.d. | 47° | n.d. | ||
| Soretii | +0.22 | n.d. | 61° | n.d. | |||
Chromophore orientations in cyt c. S is the macroscopic orientation parameter of the lipid vesicle-protein samples. Normalized LDr values and insertion angles (α) relative to the membrane normal.
S was determined from the LDr of retinoic acid as described in Materials and Methods. Retinoic acid was added after the cyt c LD spectrum had been recorded.
Fraction of bound cyt c was taken from Table 1.
LDr values given are the average LDr of La(trp) and the Lb(tyr) transitions in the four tyrosines. The relative contribution from each transition could not be resolved.
The spectral differences in the 220–230 nm region support the idea that reduction of cyt c results in rearrangement of one or several of the aromatic side chains. Both tryptophan and tyrosine display relatively strong absorption in this region due to the Bb(trp) and La(tyr) transitions, respectively. These absorption bands have similar (nearly Gauss-formed) shapes and in addition there is overlap with the peptide bond n→π∗ transitions. Therefore, the spectral change in this region cannot be quantified, and instead is described qualitatively. Fig. 2 B shows that the oxidized cyt c spectrum has a narrow positive peak at ∼230 nm that is absent in the reduced cyt c spectrum. The negative peak at ∼225 nm is larger in the reduced spectrum compared to the oxidized spectrum and the peak is displaced a few nanometers to the red. The spectral change is in agreement with the notion that either the Bb(trp) transition or the average La(tyr) transition changes sign from positive to negative upon reduction of cyt c.
LD of cyt c—effects of membrane surface charge density and protein/lipid ratio
Fig. 4 shows LD spectra of oxidized (A) and reduced (B) cyt c in lipid bilayers composed of POPC/POPG with respectively 20, 40, and 60 mol % POPG (negatively charged lipid). All spectra in each set (Fig. 4, A and B) display very similar signatures, but there is a variation in LD magnitudes that is particularly evident in the stronger absorption bands. It is apparent that the highest degree of orientation is obtained when cyt c is bound to POPC/POPG membranes with a 40% negative charge (black lines). The overall similarity suggests that the orientation of membrane-bound cyt c is essentially independent or only marginally affected by the surface charge density of the lipid membrane.
It has been suggested that the binding geometry of cyt c can vary with the protein/lipid ratio (14). We performed an incremental titration series covering protein/lipid ratios from 1:200 up to 1:18 to explore possible variations in binding mode. However, all recorded spectra had virtually identical shapes (spectra not shown).
LD of cyt c—interactions with CL
Cyt c has been reported to interact in a specific manner with the doubly negatively charged lipid CL (42–44). We tested how this lipid affects cyt c LD by incorporating CL into POPC lipid vesicles at two different molar ratios (10 or 20 mol % CL, corresponding to 20 and 40 mol % negative charge, respectively). Fig. 5 shows LD spectra of oxidized and reduced cyt c associated to POPC/CL lipid vesicles containing 20 mol % CL, a level that resembles the actual physiological level in the inner mitochondrial membrane (38). The LD spectra in Fig. 5 differs substantially from those shown in Figs. 2 B and 4, indicating the existence of a specific interaction between cyt c and CL that is not observed in presence of POPG. The differences are further described below but were also quantified in terms of orientation of the Lb transition moment in Trp59 and the x- and y-Soret transitions in the heme group using TEM analysis (see Table 2). In contrast to the spectra in Fig. 5, incorporating a smaller amount of CL into the lipid vesicles (10 mol %) had no major effect on the recorded LD (data not shown).
The overall magnitudes of the absorption bands in Fig. 5 are significantly smaller than those in Figs. 3 and 4. This cannot be ascribed to a lower degree of macroscopic orientation in these samples since the spectra in Figs. 3 B, 4, and 5 were all normalized with respect to the orientation parameter (S). Moreover, as can be seen in Table 2, the orientation is better in samples with POPC/CL vesicles than in samples with POPC/POPG vesicles (S is higher), indicating that the poor orientation of cyt c is due to the fact that the protein itself does not align so well at the membrane surface when CL is present.
For oxidized cyt c, the main changes in LD resulting from exchanging 40 mol % POPG with 20 mol % CL (the same surface charge) includes a halving of the negative Soret peak at ∼415 nm, a radical increase in the positive LD at 230 nm and a concomitant reduction of the negative LD at 225 nm. For reduced cyt c the main changes include the appearance of a bisignate negative absorption band in the Soret region, a strongly negative contribution to LD in the aromatic region, with a negative contribution from Lb(trp), and the appearance of a split signal in the 220–230 nm region resembling that observed for oxidized cyt c. The orientation of the α-helices in both oxidized and reduced cyt c bound to CL-containing lipid membranes are still on average parallel to the membrane surface, as evidenced by the positive LD at 205–210 nm, suggesting some resemblance between the different conformations found in CL and POPC/POPG membranes.
Structural models for oxidized and reduced cyt c bound to POPC/POPG lipid membranes
The angular information in Table 2 was used to construct structural models of the cyt c binding geometry in POPC/POPG lipid membranes according to the procedures described in the Materials and Methods section. For oxidized cyt c the coordinates in the 1AKK .pdb file (describing a solution NMR structure of the oxidized protein) had to be rotated around the z and x axes by the angles −76.6° and 24.1°, respectively, to fit the transition moment x,y,z coordinates with the experimental LDr values for the Lb(trp) and the resultant heme Soret transition (see Materials and Methods). In the fitted model the Lb(trp) transition moment adopted an angle to the z axis of 94° (this equals 86° because of the uniaxial distribution around the z axis) and the angle between the z axis and the normal of the heme plane was 44°. For reduced cyt c the coordinates in the 1GIW .pdb file (describing a solution NMR structure of the reduced protein) were rotated 212.5° and 27.4° around the z and x axis, respectively. In the fitted model the Lb transition moment angle to the z axis was calculated to 94° and the angle between the z axis and the heme plane normal was 48°.
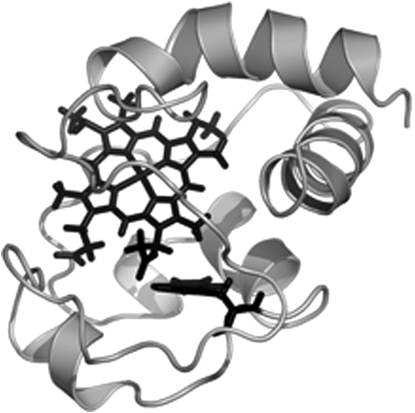
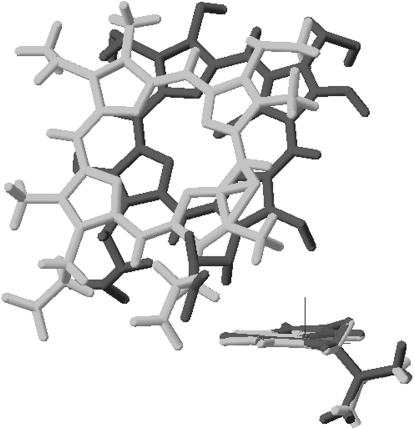
Fig. 6 shows the proposed protein orientation for the reduced state of cyt c. Fig. 7 shows an overlay of Trp59 and the heme group in reduced and oxidized cyt c and highlights the identical orientation of the indole side chain in both states, as well as the minor displacement of the heme group. It is obvious from this figure, as well as from Fig. 6, that the indole in tryptophan is oriented almost perfectly parallel to the surface of the lipid bilayer. This conveys that the orientation of the La(trp) and Bb(trp) transition moments must be close to 90°. This gives some clues as to the average orientation of tyrosines in the oxidized and reduced state, which we will return to in the Discussion.
Figure 6.
Model of reduced cyt c associated to POPC/POPG (60:40) lipid vesicles. The membrane normal is vertical in the picture and the lipid bilayer is at the bottom. The heme group and the tryptophans are colored in black.
Figure 7.
Superimposition of the membrane-binding models for oxidized (light gray) and reduced (dark gray) cyt c showing Trp59 and the heme group.
Discussion
In this study we aimed to gain insight into the potential of a new approach for assigning the binding geometry of membrane-bound proteins. We demonstrate that polarized light spectroscopy (LD) can successfully be combined with data from an atomic-resolution NMR structure obtained in solution to provide information about the binding geometry and orientation of a membrane-associated protein, here exemplified by cyt c. We further show how small structural changes within the protein can be identified, and that the methodology is sensitive enough to pick up on small differences in binding geometry between two isoforms of the same protein.
In brief, our results can be summarized as follows:
-
1.
We have demonstrated that cyt c binds to POPC/POPG lipid vesicles with a high degree of orientation, indicating the existence of only one relevant binding mode within the investigated concentration regime.
-
2.
We have shown that cyt c undergoes some minor changes in binding geometry while changing its redox state, but the average orientation of its peptide backbone (foremost the α-helical segments) is practically unchanged.
-
3.
We constructed a binding model for cyt c based on LD data and existing atomic-resolution NMR solution structures that shows how the protein binds to POPC/POPG lipid vesicles, with its α-helices on average parallel to the membrane surface, the plane of the Trp59 side chain (indole) almost perfectly perpendicular to the membrane normal, and the heme group in a tilted orientation in which the normal of the porphyrin plane makes an angle of ∼45° to the membrane normal (see Figs. 6 and 7). The model is in good agreement with earlier constraints on protein orientation suggested in the literature.
-
4.
We confirmed the existence of a particular interaction of cyt c with CL, and the combined LD and CD data point to a partial unfolding of the protein resulting in changed secondary structure and reorientation of both Trp59 and the heme group.
Obtaining angular information on the orientation of specific transition moments from LD spectra is meaningful only if it can be validated that the molecule in question adopts one binding mode. In the case of poor alignment or multiple binding modes, the LD will inevitably be reduced due to averaging effects, and consequently all computed insertion angles will approach 54.7° (magic angle, LD = 0). Therefore, it was important to justify the existence of one single binding mode for cyt c and to verify that the protein was well aligned within the membrane. Several features of this study, each of which will be discussed and justified below, indicate that this holds true for cyt c associated to lipid vesicle membranes. The mere existence of LD from samples containing cyt c and lipid vesicles shows that some preferred orientation exists and proves that the protein does not simply adsorb onto the bilayer in a random fashion. Inspection of the LD and absorption spectra in Fig. 2 B reveals that cyt c gives rise to sharp LD bands that parallel the shape of the corresponding absorption bands. This is an indication of one binding mode, since multiple binding modes often result in absorption broadening due to the heterogeneous environment around each chromophore, as well as LD shifts resulting from the fact that all chromophores are not oriented in the same fashion. Most of the estimated LDr values and corresponding insertion angles (see Table 2) are far from magic angle conditions, and the fact the orientation of Lb(trp) approaches 90° is strong evidence for that multiple orientations of at least Trp59 do not exist. This indicates that cyt c is extremely well aligned; in fact, its degree of orientation is comparable to that of retinoic acid (11), which we routinely use to sense the overall orientation of the lipid vesicles.
Although we were only able to find one binding mode for cyt c, other studies have indicated as many as three different binding models: a peripheral binding mode, an integral membrane insertion mode, and a “protein/protein interactive” binding mode (14). The LD technique cannot give information on the degree of protein insertion into the membrane, and we cannot fully distinguish between the peripheral and integral binding modes. However, most experiments in this study were performed at a protein/lipid ratio of 1:200, which should give the peripheral binding mode. According to Oellerich et al. (14), the partially inserted mode should occur at high protein/lipid ratios (above 1:18), but, as mentioned above, the appearance of a different binding mode under these conditions could not be detected by us. The “protein/protein interactive” binding mode would have given rise to exciton coupling effects (12), which were not observed. Therefore, to conclude, we believe that the peripheral binding mode is the only binding mode observed in this study.
Our method for constructing an “atomic-resolution” binding model of cyt c relies on the fact that it is possible to dock a protein structure obtained in solution onto a model lipid membrane with the help of constraints on its orientation obtained from the LD of a small number of transition moments within the protein. Therefore, it is very important that one can assume that the protein retains, to a significant extent, its tertiary structure in the membrane-bound state. The CD data presented in Fig. 1 and Table 1 indicate that the secondary structure of cyt c is similar in solution and in a POPC/POPG lipid membrane. The estimated α-helical contents (30–40%) are in good agreement with data presented by Banci et al. (22,23), and the secondary structure analysis of cyt c CD spectra (Table 1) confirms that only minor (<10%) changes in α-helical content occur upon binding to POPC/POPG lipid bilayers. Additionally, the cyt c secondary structure is essentially preserved after changes in the protein redox state, both in solution and in POPC/POPG lipid membranes. In addition, the CD spectra of bound and free protein were always similar in the Soret band. This is in contrast to the findings of Oellerich et al. (14), who observed a major difference in Soret CD when cyt c interacted with lipid vesicles composed of anionic DOPG. Both Oellerich et al. (14) and Pinheiro et al. (45) previously interpreted the change in this split CD signal in the Soret absorption band of cyt c as a disruption of the tight packing of core residues in cyt c upon binding to the lipid membrane. The absence of such a change in Fig. 1 indicates that the protein environment near the heme group is essentially unaffected by lipid interactions under the experimental conditions used in our study. This holds true for all samples prepared during this study, since the effect was similar throughout. Therefore, we believe it is reasonable to assume that cyt c retains its tertiary structure upon interaction with POPC/POPG lipid membranes.
A binding model of cyt c bound to POPC/POPG membranes (Fig. 6) was constructed using two orientational constraints: the direction of the Lb(trp) transition moment and the direction of the normal to the heme plane. It would of course have been desirable to assign, with certainty, more transition moment directions and include them to produce a refined model of membrane-bound cyt c. This could have been achieved by employing the site-specific LD by molecular replacement (SSLD-MR) approach, in which one aromatic amino acid at a time is mutated and the resulting LD spectrum is compared against the wild-type protein to obtain the LD of that particular residue (46,47). This approach would have made it possible to assign, for example, the orientation of each individual tyrosine, albeit with a significantly higher experimental expenditure. Here, we will instead concentrate on demonstrating the power of the LD technique in its simplest form by showing that several LD features that were not included to create the current model can in fact be remarkably well predicted from the theoretical orientation of transition moments in the model.
All LD spectra of cyt c in POPC/POPG lipid vesicles (see Figs. 2 B and 4) display positive peaks at 208–210 nm, showing that cyt c must be oriented with its α-helices more parallel than perpendicular to the membrane surface. The magnitude of the LD in this region is also a good indication that the α-helices should on average make a large angle to the membrane normal. These features are confirmed in the model where the two longest helical segments, corresponding to residues 3–12 and 90–98, are oriented almost parallel to the helix surface. The fact that the LD at 210 nm remains constant upon reduction of cyt c suggests that the overall orientation of the protein does not change upon reduction. This is also the case in our model (not shown).
According to Figs. 6 and 7, our models predict that Trp59 is oriented with the indole plane parallel to the surface of the lipid membrane in both oxidized and reduced cyt c. This infers that not only the Lb(trp) but also the La(trp) and Bb(trp) transition moments are close to parallel to the surface of the lipid membrane and should thus exhibit strongly positive LD. For La(trp) this should amount to an LDr in parity with that of Lb(trp) (+0.74; see Table 2). If the Lb(trp) contribution is subtracted from the LD spectrum, the residual LDr is +0.30 for the oxidized form and zero for the reduced form. Therefore, we can expect that the Lb transitions in the four tyrosine residues of cyt c should on average display a net negative LD in the 250–300 nm region. By representing each Lb(tyr) with a vector in the model in Fig. 6 (and in the corresponding model of oxidized cyt c), we calculated their angles relative to the z axis and then computed their theoretical LDr values. This calculation showed that the net LDr for Lb(tyr) should indeed be negative in both oxidized and reduced cyt c. Moreover, the theoretical net LDr for Lb(tyr) is more negative in reduced cyt c compared to the oxidized form, which explains the observed change in LD upon reduction of cyt c. We can therefore conclude, from LD data and with the help of the atomic-resolution data of the rotated NMR structures that were used to build our models of membrane-bound cyt c, that although Trp59 is unaffected by the redox state of cyt c, one or several of the four tyrosines do change orientation.
The appearance of a split LD signal in the Soret band indicates immediately that the heme group should be tilted with respect to the membrane normal. This bisignate LD in the Soret band was observed before by our group (48) and others (32). Rajendra et al. (32) calculated a tilt angle between the heme plane and the average lipid of ∼50°, which is in agreement with what we observed. The fact that both the oxidized and reduced cyt c gives rise to opposite LD signatures in the Soret band indicates at first glance a significant rearrangement of the heme when the protein changes redox state. However, there is only a marginal difference in the actual orientation of the heme normal (whose LDr was computed from the LDr of the x,y-Soret transitions). The similarity in orientation of the heme group is emphasized in the cartoon model in Fig. 7. The heme normal is 44° from the membrane normal in the oxidized form and 48° in the reduced form. This minor change in tilt is probably not relevant for the function of cyt c and shall not be further discussed here. The shift of signs in the Soret band is instead assigned to a change in the local perturbation of the near-degenerate transitions of the Soret band.
It has been suggested that cyt c contains a specific lipid-binding site consisting of a region of clustered positive charges (14). Characteristically, most of the basic residues in cyt c are segregated into well-defined patches on the surface of the protein molecule (16). Peripheral binding of the protein to negatively charged phosphatidyl headgroups has been attributed to lysine-rich domains around the exposed heme edge on the surface of the protein (50,51). In particular, it has been suggested that the four lysines (72, 73, 86, and 87) should be oriented on the membrane-facing side of cyt c (52–55). Our model of membrane-bound cyt c (Fig. 6) places Lys72 and Lys73 on the bottom half of the spherical protein. By contrast, Lys86 and Lys87 are located on the upper half of the protein, which disagrees with the notion that they are part of a lipid-binding site. For these residues to be in close contact with negative moieties in the lipid membrane, cyt c must be deeply inserted, which is not very likely given the polar character of the protein. It should be emphasized that it is not possible to rotate the protein so that all four tyrosines are placed on the lower half of cyt c if its α-helical fragments are to be well aligned with the lipid surface. Therefore, it seems that not all lysines are important for lipid interaction in the peripheral binding mode. Vanderkooi and co-workers (56) suggested that cyt c binds to the inner mitochondrial membrane on the side of the protein where Met65 is located. Such an orientation was also indicated by Brown and Wuthrich (57). Our model (see Fig. 6) places Met65 at the lower hemisphere, in good agreement with those observations.
It has been proposed that cyt c interacts specifically with CL, and two different binding sites for CL on cyt c have been suggested (42–44). In this study we found that the LD of cyt c is clearly affected by the presence of CL, but we also observed, somewhat surprisingly, that the cyt c binding affinity for lipid vesicles containing this lipid is lower than the affinity for POPC/POPG vesicles with identical surface charge (see Table 1). In addition, we observed a much smaller LD signal from cyt c upon CL incorporation, indicating poorer alignment within the membrane. The overall lower α-helicity of cyt c associated to CL vesicles suggests that the protein partially unfolds upon interaction with CL lipids, and is in agreement with previous observations that CL may induce loosening of the cyt c tertiary structure (52,58). No significant changes could however be observed in the Soret region of the CD spectrum. The unfolding itself may result in cyt c losing some of its orientation specificity, which would explain the lower overall LD signals in Fig. 5. However, there are also some spectral changes in the aromatic region and the Soret region that indicate reorientation of these residues upon interaction with CL. For oxidized cyt c, this change is mainly observed in the aromatic spectral region. The insertion angle for Lb(trp) is 72°.
The LD of the reduced state of cyt c is more drastically altered by interaction with CL. In the aromatic region, the LD curve indicates that a major reorientation of Trp59 has occurred because the characteristic “double peak” of the Lb(trp) transition has changed sign and is negative, as is the residual LD describing the average orientation of La(trp) and Lb(tyr) transition moments. There is still a positive peak at ∼230 nm, which is larger than for the spectra in Fig. 2 B, but the magnitude is much smaller than in the oxidized sample. This suggests that upon interaction with CL, Trp59 in reduced cyt c is significantly tilted toward the membrane normal, and therefore we can also conclude that the interaction with CL affects the structure of the protein in the region where Trp59 is positioned. Since Trp59 is buried within the core of cyt c in its native water-soluble state, this could indicate a major rearrangement of segments in the protein, and may be compatible with the finding that cyt c contains a binding pocket for lipid acyl chains (44). The secondary structure content of reduced cyt c in CL-containing vesicles (20% CL) is fairly similar to what is observed in POPC/POPG, but one may speculate about a certain loosening of the protein tertiary structure rather than an unfolding. This loosening likely retains Trp59 within the protein, albeit with a changed orientation. Further, reduced cyt c gives rise to a peculiar LD in the Soret band signified by two well-separated negative peaks. This may be interpreted to mean that CL comes in close contact with the heme group and decreases the degeneracy of the x,y-Soret transitions (which become further apart in energy and thus also in wavelength).
It is clear that a particular type of interaction occurs between CL and cyt c. Our data also indicate that this particular interaction can disturb the structure and binding geometry of the entire protein, and that it brings about a pronounced difference between the oxidized and reduced states that is not observed in the POPC/POPG lipid system.
Conclusions
We have shown what we consider a novel approach to assign the binding geometry of membrane-bound proteins by combining polarized spectroscopy with NMR structures. The possibility of obtaining biological relevant structural information about membrane-associated proteins with the proteins situated in their normal surroundings has great potential. This fairly simple experimental procedure can be used to obtain detailed structural data, and may be useful for monitoring in situ small structural changes, such as the effects of redox state variations or interactions with membrane constituents.
Acknowledgments
This work was funded by grants to B. Nordén from the Swedish Cancer Foundation (Cancerfonden) and the European Commission's Sixth and Seventh Framework Programmes (AMNA contract 013575 and ZNIP contract 037783), and by a grant to E. K. Esbjörner from the Lennander Foundation.
Contributor Information
Christina E.B. Caesar, Email: christina.caesar@chalmers.se.
Elin K. Esbjörner, Email: eline@chalmers.se.
References
- 1.Alberts B., Johnson A., Walter P., Lewis J., Raff M. Taylor & Francis; London, UK: 2008. Molecular Biology of the Cell. [Google Scholar]
- 2.Drews J. Drug discovery: a historical perspective. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 3. http://www.rcsb.org/pdb/home/home.do
- 4.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N. The Protein Data Bank. Nucl. Acid. Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Opella S.J. NMR and membrane proteins. Nat. Struct. Biol. 1997;4(Suppl):845–848. [PubMed] [Google Scholar]
- 6.Ardhammar M., Mikati N., Norden B. Chromophore orientation in liposome membranes probed with flow dichroism. J. Am. Chem. Soc. 1998;120:9957–9958. [Google Scholar]
- 7.Ardhammar M., Lincoln P., Nordén B. Ligand substituents of ruthenium dipyridophenazine complexes sensitively determine orientation in liposome membrane. J. Phys. Chem. 2001;105:11363–11368. [Google Scholar]
- 8.Ardhammar M., Lincoln P., Nordén B. Invisible liposomes: refractive index matching with sucrose enables flow dichorism assesment of peptide orientation in lipid vesicle membrane. Proc. Natl. Acad. Sci. USA. 2002;99:15313–15317. doi: 10.1073/pnas.192583499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brattwall C.E.B., Lincoln P., Norden B. Orientation and conformation of cell-penetrating peptide penetratin in phospholipid vesicle membranes determined by polarized-light spectroscopy. J. Am. Chem. Soc. 2003;125:14214–14215. doi: 10.1021/ja0366989. [DOI] [PubMed] [Google Scholar]
- 10.Caesar C.E., Esbjorner E.K., Lincoln P., Norden B. Membrane interactions of cell-penetrating peptides probed by tryptophan fluorescence and dichroism techniques: correlations of structure to cellular uptake. Biochemistry. 2006;45:7682–7692. doi: 10.1021/bi052095t. [DOI] [PubMed] [Google Scholar]
- 11.Svensson F.R., Lincoln P., Norden B., Esbjorner E.K. Retinoid chromophores as probes of membrane lipid order. J. Phys. Chem. B. 2007;111:10839–10848. doi: 10.1021/jp072890b. [DOI] [PubMed] [Google Scholar]
- 12.Esbjorner E.K., Caesar C.E.B., Albinsson B., Lincoln P., Norden B. Tryptophan orientation in model lipid membranes. Biochem. Biophys. Res. Commun. 2007;361:645–650. doi: 10.1016/j.bbrc.2007.07.135. [DOI] [PubMed] [Google Scholar]
- 13.Esbjorner E.K., Oglecka K., Lincoln P., Graslund A., Norden B. Membrane binding of pH-sensitive influenza fusion peptides. positioning, configuration, and induced leakage in a lipid vesicle model. Biochemistry. 2007;46:13490–13504. doi: 10.1021/bi701075y. [DOI] [PubMed] [Google Scholar]
- 14.Oellerich S., Lecomte S., Paternostre M., Heimburg T., Hildebrandt P. Peripheral and integral binding of cytochrome c to phospholipids vesicles. J. Phys. Chem. B. 2004;108:3871–3878. [Google Scholar]
- 15.Chen Y.H., Yang J.T., Chau K.H. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 16.Dickerson R.E., Takano T., Eisenberg D., Kallai O.B., Samson L. Ferricytochrome c. I. General features of the horse and bonito proteins at 2.8 A resolution. J. Biol. Chem. 1971;246:1511–1535. [PubMed] [Google Scholar]
- 17.Kalanxhi E., Wallace C.J.A. Cytochrome c impaled: investigation of the extended lipid anchorage of a soluble protein to mitochondrial membrane models. Biochem. J. 2007;407:179–187. doi: 10.1042/BJ20070459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orrenius S. Mitochondrial regulation of apoptotic cell death. Toxicol. Lett. 2004;149:19–23. doi: 10.1016/j.toxlet.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Teissie J. Interaction of cytochrome c with phospholipid monolayers. Orientation and penetration of protein as functions of the packing density of film, nature of the phospholipids, and ionic content of the aqueous phase. Biochemistry. 1981;20:1554–1560. doi: 10.1021/bi00509a023. [DOI] [PubMed] [Google Scholar]
- 20.Pinheiro T.J.T. The interaction of horse heart cytochrome c with phospholipid bilayers. Structural and dynamic effects. Biochimie. 1994;76:489–500. doi: 10.1016/0300-9084(94)90173-2. [DOI] [PubMed] [Google Scholar]
- 21.Domanov Y.A., Molotkovsky J.G., Gorbenko G.P. Coverage-dependent changes of cytochrome c transverse location in phospholipid membranes revealed by FRET. Biochim. Biophys. Acta. 2005;1716:49–58. doi: 10.1016/j.bbamem.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Banci L., Bertini I., Gray H.B., Luchinat C., Reddig T. Solution structure of oxidized horse heart cytochrome. c. Biochemistry. 1997;36:9867–9877. doi: 10.1021/bi970724w. [DOI] [PubMed] [Google Scholar]
- 23.Banci L., Bertini I., Huber J.G., Spyroulias G.A., Turano P. Solution structure of reduced horse heart cytochrome. c. J. Biol. Inorg. Chem. 1999;4:21–31. doi: 10.1007/s007750050285. [DOI] [PubMed] [Google Scholar]
- 24.Qi P.X., Beckman R.A., Wand A.J. Solution structure of horse heart ferricytochrome c and detection of redox-related structural changes by high-resolution 1H NMR. Biochemistry. 1996;35:12275–12286. doi: 10.1021/bi961042w. [DOI] [PubMed] [Google Scholar]
- 25.Heimburg T., Marsh D. Protein surface-distribution and protein-protein interactions in the bining of pheripheral proteins to charged lipid membranes. Biophys. J. 1995;68:536–546. doi: 10.1016/S0006-3495(95)80215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sreerama N., Woody R.W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 27.Manavalan P., Johnson W.C. Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem. 1987;167:76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 28.Compton L.A., Johnson W.C. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 29.Whitmore L., Wallace B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32(Web Server issue):W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitmore L., Wallace B.A. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 31.Nordén B., Kubista M., Kurucsev T. Linear dichroism spectroscopy of nucleic acids. Q. Rev. Biophys. 1992;25:51–170. doi: 10.1017/s0033583500004728. [DOI] [PubMed] [Google Scholar]
- 32.Rajendra J., Damianoglou A., Hicks M., Booth P., Rodger P.M. Quantitation of protein orientation in flow-oriented unilamellar liposomes by linear dichroism. Chem. Phys. 2006;326:210–220. [Google Scholar]
- 33.Eaton W.A., Hochstrasser R.M. Electronic spectrum of single crystals of ferricytochrome-c. J. Chem. Phys. 1967;46:2533–2539. doi: 10.1063/1.1841081. [DOI] [PubMed] [Google Scholar]
- 34.Gouterman M. The Porphyrins. In: Dolphin D., editor. Academic Press; New York: 1978. pp. 1–167. [Google Scholar]
- 35.Adar F. The Porphyrins. In: Dolphin D., editor. Academic Press; New York: 1978. pp. 167–209. [Google Scholar]
- 36.Thulstrup E.W., Michl J., Eggers J.H. Polarization spectra in stretched polymer sheets. II. Separation of π-π∗ absorption of symmetrical molecules into components. J. Phys. Chem. 1970;74:3868–3878. [Google Scholar]
- 37.Albinsson B., Norden B. Excited-state properties of the indole chromophore: electronic transition moment directions from linear dichroism measurements: effect of methyl and methoxy substituents. J. Phys. Chem. 1992;96:6204–6212. [Google Scholar]
- 38.Cronier F., Patenaude A., Gaudreault R.C., Auger M. Membrane composition modulates the interaction between a new class of antineoplastic agents deriving from aromatic 2-chloroethylureas and lipid bilayers: a solid-state NMR study. Chem. Phys. Lipids. 2007;146:125–135. doi: 10.1016/j.chemphyslip.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Sanishvili R., Volz K.W., Westbrook E.M., Margoliash E. The low ionic strength crystal structure of horse cytochrome c at 2.1 A resolution and comparison with its high ionic strength counterpart. Structure. 1995;3:707–716. doi: 10.1016/s0969-2126(01)00205-2. [DOI] [PubMed] [Google Scholar]
- 40.Moffitt W., Yang J.T. The optical rotatory dispersion of simple polypeptides. I. Proc. Natl. Acad. Sci. USA. 1956;42:596–603. doi: 10.1073/pnas.42.9.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moffitt W. The optical rotatory dispersion of simple polypeptides. II. Proc. Natl. Acad. Sci. USA. 1956;42:736–746. doi: 10.1073/pnas.42.10.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trumpower B.L., Gennis R.B. Energy transduction by cytochrome complexes in mitochondrial and bacterial respiration: the enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annu. Rev. Biochem. 1994;63:675–716. doi: 10.1146/annurev.bi.63.070194.003331. [DOI] [PubMed] [Google Scholar]
- 43.Gorbenko G.P., Molotkovsky J.G., Kinnunen P.K.J. Cytochrome c interaction with cardiolipin/phosphatidylcholine model membranes: effect of cardiolipin protonation. Biophys. J. 2006;90:4093–4103. doi: 10.1529/biophysj.105.080150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rytomaa M., Kinnunen P.K. Evidence for two distinct acidic phospholipid-binding sites in cytochrome. c. J. Biol. Chem. 1994;269:1770–1774. [PubMed] [Google Scholar]
- 45.Pinheiro T.J., Elöve G.A., Watts R., Roder H. Structural and kinetic description of cytochrome c unfolding induced by the interaction with lipid vesicles. Biochemistry. 1997;36:13122–13132. doi: 10.1021/bi971235z. [DOI] [PubMed] [Google Scholar]
- 46.Frykholm K., Morimatsu K., Norden B. Conserved conformation of RecA protein after executing the DNA strand-exchange reaction. A site-specific linear dichroism structure study. Biochemistry. 2006;45:11172–11178. doi: 10.1021/bi060621q. [DOI] [PubMed] [Google Scholar]
- 47.Morimatsu K., Takahashi M., Norden B. Arrangement of RecA protein in its active filament determined by polarized-light spectroscopy. Proc. Natl. Acad. Sci. USA. 2002;99:11688–11693. doi: 10.1073/pnas.142404499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodger A., Rajendra J., Marrington R., Ardhammar M., Nordén B. Flow oriented linear dichroism to probe protein orientation in membrane environments. Phys. Chem. Chem. Phys. 2002;4:4051–4057. [Google Scholar]
- 49.Reference deleted in proof.
- 50.Salamon Z., Tollin G. Surface plasmon resonance studies of complex formation between cytochrome c and bovine cytochrome c oxidase incorporated into a supported planar lipid bilayer. I. Binding of cytochrome c to cardiolipin/phosphatidylcholine membranes in the absence of oxidase. Biophys. J. 1996;71:848–857. doi: 10.1016/S0006-3495(96)79286-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szebeni J., Tollin G. Interaction of cytochrome c with liposomes: covalent labeling of externally bound protein by the fluorescent probe, azidonaphthalenedisulfonic acid, enclosed in the inner aqueous compartment of unilamellar vesicles. Biochim. Biophys. Acta. 1988;932:153–159. doi: 10.1016/0005-2728(88)90150-8. [DOI] [PubMed] [Google Scholar]
- 52.Spooner P.J.R., Watts A. Cytochtome c interactions with cardiolipin in bilayers: a multinuclear magic-angle spinning NMR study. Biochemistry. 1991;30:3871–3879. doi: 10.1021/bi00156a037. [DOI] [PubMed] [Google Scholar]
- 53.Pinheiro T.J. The interaction of horse heart cytochrome c with phopholipid bilayers. Structural and dynamic effects. Biochimie. 1994;76:489–500. doi: 10.1016/0300-9084(94)90173-2. [DOI] [PubMed] [Google Scholar]
- 54.Gorbenko G.P. Structure of cytochrome c complexes with phospholipids as revealed by resonance energy transfer. Biochim. Biophys. Acta. 1999;1420:1–13. doi: 10.1016/s0005-2736(99)00082-6. [DOI] [PubMed] [Google Scholar]
- 55.Kostrezewa A., Pali T., Froncisz W., Marsh D. Membrane location of spin-labeled cytochrome c determined by paramagnetic relaxation agents. Biochemistry. 2000;30:6066–6074. doi: 10.1021/bi992559l. [DOI] [PubMed] [Google Scholar]
- 56.Vanderkooi J., Erecinska M., Chance B. Cytochrome c interaction with membranes. II. Comparative study of the interaction of c cytochromes with the mitochondrial membrane. Arch. Biochem. Biophys. 1973;157:531–540. doi: 10.1016/0003-9861(73)90672-3. [DOI] [PubMed] [Google Scholar]
- 57.Brown L.R., Wuthrich K. NMR and ESR studies of the interactions of cytochrome c with mixed cardiolipin-phosphatidylcholine vesicles. Biochim.Biophys. Acta. 1977;468:389–410. doi: 10.1016/0005-2736(77)90290-5. [DOI] [PubMed] [Google Scholar]
- 58.Sankaram M.B., de Kruijff B., Marsh D. Selectivity of interaction of spin-labelled lipids with peripheral proteins bound to dimyristoylphosphatidylglycerol bilayers, as determined by ESR spectroscopy. Biochim. Biophys. Acta. 1989;986:315–320. [Google Scholar]
- 59.Lakowicz J.R. Springer; New York: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]