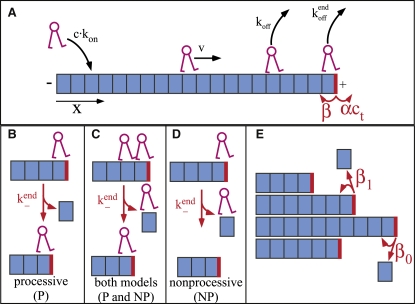
Figure 1.
Model of kinesin-8 motor protein's interaction with a MT protofilament showing the key rates. Rates in black affect only the motor, while those in red affect the MT plus-end. The plus-end of the MT protofilament is indicated by a thick vertical (red) line, and the dimers are indicated by (blue) boxes. Note that depolymerization (at rate k−end) affects both the motor and the MT plus-end. (A) A motor binds to a dimer of the MT with on rate konc and unbinds with rate koff. The motor steps forward at rate v; backward motion is not considered, due to the biased motion of kinesin 8s. MT dynamics are represented by allowing dimers to add to a MT-end at rate αct (where ct is the bulk concentration of tubulin dimers) and dissociate at rate β. (B–D) Depolymerization models. (B) If the motor depolymerizes processively, it removes a MT dimer as it steps backward (with rate k−end), thereby shortening the MT. (C) If the dimer behind the MT-end is occupied, the motor falls off the MT in either model. (D) In the nonprocessive depolymerization model, the motor removes a single tubulin dimer and falls off the MT. (E) Lateral interactions help stabilize MTs. We incorporated this into our model by allowing the depolymerization rate to depend on the number of neighboring protofilaments (i.e., 0, 1, or 2). In this case, the rate at which a terminal tubulin dimer unbinds from a protofilament is given by β0 if the dimer has no lateral neighbors; β1 if the dimer has one lateral neighbor; and β2 if the dimer has two lateral neighbors.