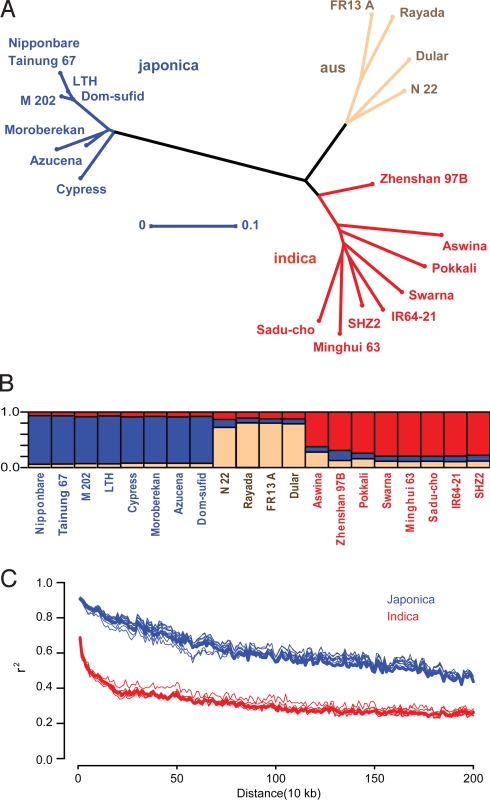
Fig. 2.
Phylogenetic relationships, population structure, and decay of LD in the OryzaSNPset. (A) Unweighted neighbor-joining dendrogram for nonrepetitive SNPs in the MBML-intersect data (159,879 sites). Horizontal bar indicates distance by simple matching coefficient. (B) Population structure as determined by MB inference using InStruct (24). The 3 groups correspond to indica (red), aus (brown), and japonica (blue). (C) Decay of LD, expressed as r2 as a function of inter-SNP distance for filtered MBML-intersect SNPs, in the indica and japonica varieties, for each chromosome (light) and overall (bold). Limited numbers of japonica SNPs bias LD estimates.