Table 1.
Performance comparison of sampling method and guided halving algorithm in the case of one outgroup
| Halving analysis |
Sampling method |
Guided halving |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R−T | 2n | dt,x⊕x | dx,r | dmin | da,r | ΔA | da,a* | Time | dx,r | da,r | ΔA | 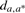 |
Time |
| AG-CG | 538 | 186 | 204 | 196 | 180 | −16 | 156 | 37 | 153 | 153 | 0 | 120 | 2.3 |
| AG-SC | 1012 | 119 | 237 | 229 | 208 | −21 | 53 | 158 | 184 | 183 | −1 | 32 | 5.3 |
| KL-CG | 546 | 186 | 210 | 203 | 184 | −19 | 154 | 50 | 160 | 160 | 0 | 120 | 3.5 |
| KL-SC | 1026 | 122 | 241 | 232 | 216 | −16 | 51 | 140 | 197 | 197 | 0 | 39 | 6.1 |
| KW-CG | 542 | 188 | 247 | 238 | 230 | −8 | 167 | 26 | 216 | 215 | −1 | 142 | 3.3 |
| KW-SC | 994 | 121 | 364 | 355 | 350 | −5 | 70 | 72 | 325 | 323 | −2 | 41 | 5.1 |
| A*-CG | 600 | 199 | 183 | 169 | 129 | −40 | 129 | 81 | 84 | 84 | 0 | 84 | 1.5 |
| A*-SC | 1062 | 124 | 79 | 70 | 37 | −33 | 37 | 114 | 5 | 5 | 0 | 5 | 0.3 |
| AG-V | 576 | 61 | 157 | 151 | 149 | −2 | 54 | 12 | 148 | 148 | 0 | 51 | 0.9 |
| KL-V | 584 | 62 | 167 | 160 | 158 | −2 | 53 | 12 | 157 | 157 | 0 | 51 | 0.9 |
| KW-V | 582 | 62 | 224 | 218 | 215 | −3 | 52 | 13 | 212 | 212 | 0 | 51 | 1.0 |
| A*-V | 600 | 62 | 57 | 49 | 39 | −10 | 39 | 14 | 29 | 29 | 0 | 29 | 0.2 |
Sample size 2000 for the sampling method. R−T represents the outgroup and doubling descendant. n is the number of genes available in that pair of genomes, with two copies in T. dt,x⊕x=d(T,X′⊕X′′) is the doubling distance, constant over all analyses. 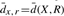 represents the average, over all samples, of the distance estimate between the ancestor, just before doubling, and the outgroup, and the adjacent entry dmin=minsampled(X,R) is the minimum found. ΔA is the improvement over d(T,X′⊕X′′)+d(X,R) due to local searching, allowing A to be found outside the set of halving solutions. da,a*=d(A, A*) is the distance between the inferred ancestor and the ‘ground truth’. Time is measured in minutes, for 2000 samples of unrestricted halving or for one GGH run.
represents the average, over all samples, of the distance estimate between the ancestor, just before doubling, and the outgroup, and the adjacent entry dmin=minsampled(X,R) is the minimum found. ΔA is the improvement over d(T,X′⊕X′′)+d(X,R) due to local searching, allowing A to be found outside the set of halving solutions. da,a*=d(A, A*) is the distance between the inferred ancestor and the ‘ground truth’. Time is measured in minutes, for 2000 samples of unrestricted halving or for one GGH run.
