Summary
Adaptation of Mycobacterium tuberculosis to an anaerobic dormant state that is tolerant to several antibacterials is mediated largely by a set of highly expressed genes controlled by DosR. A DosR mutant was constructed to investigate whether the DosR regulon is involved in antibacterial tolerance. We demonstrate that induction of the regulon is not required for drug tolerance either in vivo during a mouse infection or in vitro during anaerobic dormancy. Thus, drug tolerance observed in these models is due to other mechanisms such as the bacilli simply being in a non-replicating or low metabolic state. Our data also demonstrate that the DosR regulon is not essential for virulence during chronic murine infection. However, decreased lung pathology was observed in the DosR mutant. We also show that the DosR regulon genes are more highly conserved in environmental mycobacteria, than in pathogenic mycobacteria lacking a latent phase or environmental reservoir. It is possible that the DosR regulon could contribute to drug tolerance in human infections; however, it is not the only mechanism and not the primary mechanism for tolerance during a mouse infection. These data suggest that the regulon evolved not for pathogenesis or drug tolerance but for adaptation to anaerobic conditions in the environment and has been adapted by M. tuberculosis for survival during latent infection.
Keywords: Dormancy, DosR, Latency, Antibacterial tolerance, Cornell model
Introduction
Despite the availability of anti-tubercle drugs for five decades, M. tuberculosis continues to be one of the most prevalent and deadly infectious diseases, infecting between 25 to 43% of the world’s population and killing approximately two million people per year [1]. The difficulty in control and treatment of M. tuberculosis is not primarily due to genetic mutation, but rather to a phenotypic resistance of the bacilli to antibiotics, as bacilli that survive initial treatment with antibiotics are often fully drug-sensitive[2–4]. Neither is it access of antibiotics to the bacilli within the granuloma, as isoniazid is fully able to penetrate lesions within the human lung[5]. The mechanism(s) that confer phenotypic drug tolerance to in vivo M. tuberculosis are currently not well understood but are critical to consider for developing more effective treatment regimes.
An important aspect of M. tuberculosis which allows it to survive and thrive in the human population is the ability of the bacteria to reside in the asymptomatic host, [5, 6]. The majority of M. tuberculosis infections result in a latent infection [7]. In these latent infections, bacilli show an altered metabolic state [7–11] and undergo little if any replication as indicated by the stability of their DNA restriction fragment-length polymorphism patterns. A major challenge in studying the bacterial state during latency has been the inability of easily manipulated small rodent models to mimic the complexities of human latent infection. Therefore animal studies are often supplemented with in vitro models.
The exact stimuli which cause M. tuberculosis to enter an altered state during latent infection are uncertain. One leading model suggests that low oxygen tension and or the presence of two respiratory competitors of oxygen, nitric oxide (NO) and carbon monoxide (CO), could be cues in the host which signal or force the pathogen to adopt a non-replicating state in the absence of aerobic respiration [9, 12–15]. Adaptation to anaerobiosis has been the most widely studied in vitro model [12, 16]. During anaerobiosis M. tuberculosis ceases growth, markedly decreases RNA and protein synthesis, and enters a dormant although probably not a spore-like state[7, 11, 12]. Dormancy is used in this context to indicate a state more akin to mammalian hibernation than sporulation found in Bacillus species. Low oxygen tension, NO, or CO activate a three-component regulatory system comprised of two sensor kinases, DosS and DosT, and a response regulator, DosR[10, 14, 17, 18]. Activated DosR initiates transcription of a set of genes known as the DosR regulon, which allow the bacteria to survive long periods of anaerobiosis, and which may be important for long-term survival within the host during latent infection[8, 10, 14, 19, 20].
It has long been speculated that M. tuberculosis in a latent or dormant state may play a role in the drug tolerance observed during infection[2, 3, 5, 7, 21]. Antibacterials in use today are generally more effective against actively replicating bacteria[7, 15, 22–26]. Drug tolerance has been linked to slowed or inactive metabolism, which in the case of M. tuberculosis could be the state of the bacilli during latent infection, or of a subpopulation during active infection [8, 9, 12, 14, 27, 28]. The first-line antibiotics used to treat M. tuberculosis infection: isoniazid (INH), rifampin (RIF), pyrazinamide (PZA), and ethambutol (EMB), are all active against aerobic, actively replicating bacteria[22]. However, the effectiveness of these antibiotics is reduced or eliminated against anaerobic dormant bacilli [15, 26]. Since the DosR regulon is required for anaerobic dormancy there has been speculation that it plays a direct role in phenotypic drug tolerance [28–30].
To determine the DosR regulon’s role in M. tuberculosis drug tolerance, we constructed a dosR mutant unable to induce the entire regulon, and analyzed its survival following exposure to key antibacterials both in vivo and in vitro. To place these results in the broader context of other mycobacteria, we investigated regulon conservation across the genus.
Methods
Culture conditions and strains
Liquid cultures of M. tuberculosis strain H37Rv, the dormancy knock-out strain H37Rv:ΔRv3134c-Rv3132c (DorKO), and the complemented strain H37Rv:ΔRv3134c-3132c::Rv3134c-Rv3132c (DorCO) were maintained in Dubos-Tween-albumin broth (DTA –Difco Dubos broth base (Beckton-Dickinson), 0.5% BSA fraction V, 0.75% glucose, and 0.17% NaCl) at 37°C, and were not allowed to exceed an optical density 600 (OD600) of 0.5. The dormant cultures were maintained as previously described by Wayne and Hayes[15]. Briefly, 20 × 125 mm tubes with a headspace to culture ratio of 0.5:1 were inoculated with H37Rv, DorKO, or DorCO at an OD600 of 0.004. The tops were sealed using solid caps with rubber liners to allow the injection of antibiotics, and were stirred at 120 rpm using 8 mm stir bars.
Construction of the DorKO mutant
The three gene operon containing dosR was deleted following the protocol described by Bardarov et al[13]. Briefly, flanking regions comprising the bases upstream of Rv3134c and the downstream region of Rv3132c were amplified by PCR from M. tuberculosis H37Rv genomic DNA. For amplification of the Rv3134 upstream fragment (650bp) the following primers were employed: 34URt1, 5’-GGTACCATTAATTGGTAAGAACGCGTAGTCCA and 34UFt1, 5’-TCTAGAGTCGATACCAACGACCACTG. For amplification of the RV3132c downstream fragment (502bp) the following primers were employed: 32DRt1, 5’-AAGCTTACAGTGCTGCGATGGTCA and 32DFt1, 5’-ACTAGTATTAATGACCGACGAGGAATATCTGC. All primers were tagged with restriction sites (in bold) to aid cloning and construction of the deleted allele. The flanking regions were cloned with the pGEM-Teasy cloning kit (Promega), and sequenced. The pYUB854 plasmid was used to produce the pYUB854Rv3134c-Rv3132c::Hyg plasmid, which contains a hygromycin fragment flanked by the amplified flanking regions of Rv3134c and Rv3132c, a lambda cos site, and a unique Pac1 site. pYUB854Rv3134c-Rv3132c::Hyg was digested with Pac1, and was packaged into the unique Pac1 site of the temperature sensitive mutant of the mycobacteriophage TM4, phae87, and high titer pYUB854Rv3134c-Rv3132c::Hyg phage was prepared in M. smegmatis at 30°C. M. tuberculosis H37Rv was grown to an OD600 of 0.8, washed once with MP buffer (50 mM Tris-Cl, pH 7.4, 10mM MgSO4, 2mM CaCl2, and 150 mM NaCl), suspended in 1/10 volume of MP buffer, and infected with the phage containing pYUB854Rv3134c-Rv3132c::Hyg at an MOI of 10 for 4 hr at 37°C. Cells were spun, suspended in 7H9 media containing 0.05% Tween 80, plated on 7H10 plates containing hygromycin (50mg/ml), and incubated at 37°C for 3 weeks. Hygromycin resistant colonies were confirmed for allelic exchange using PCR and Southern analysis.
Complementation of DorKO
The dosR gene is the second gene in a three-gene operon, Rv3134c-Rv3133c(dosR)-Rv3132c(dosS). In order to construct two strains one unable to induce the DosR regulon and the other with DosR regulon regulation restored under the control of its native promoter, we deleted the entire three-gene operon. By this approach, the three-gene operon including the native upstream promoter region could be restored in the complemented strain and allows for control by possible internal regulatory and promoter elements. For complementation, the entire region from Rv3134c through Rv3132c was cloned from the MTCY3A2 cosmid (kindly provided by Dr Stewart Cole, Pasteur Institute) using the 3’ XbaI site downstream of Rv3132c and the 5’ SspI site that is 300 bases upstream of the initiation codon of Rv3134c, encompassing the operon promoter region [14]. The 4361bp fragment was cloned into unique PvuII site of the integrative shuttle vector pMVRow using the 5’ SspI site and the 3’ XbaI site, which were filled in using T4 DNA polymerase. pMVRow is a derivative of pMV361[31] with the Phsp60 promoter deleted and marked by the restriction site PacI. For constructing pMVRow inverse PCR was employed using pMV361 as a template and the high fidelity Advantage PCR mix (Clontech) with the following primers: pMVpacIfor, 5’-GGATCTTAATTAACAAGACAATTGCGGATCCAGCTG and pMVpacIrev, 5’-GGATCTTAATTAACCTCGGCCCTCCGATCCGGGTG. pMVRow containing the three-gene operon was transformed into DorKO. Hygromycin and kanamycin were used for selection of bacteria complemented for the three-gene operon.
qrt-PCR analysis of DosR regulon expression
Logarithmically growing cultures of H37Rv at an OD600 of 0.1 were exposed to nitric oxide to induce the DosR regulon by addition of the 0.1 mM of the nitric oxide donor Diethylamine NONOate (DEA-NO) for 20 min. RNA was extracted and cDNA synthesized as previously described[32]. Three separate biological replicates were obtained. The FastPCR program[17] was used to design all primers and probe sets. The primer sets and probe sequences are as follows: sigA forward (CCTACGCTACGTGGTGGATTCG), reverse (TTTGGCCAGCTCCTCGGGCGT), and probe (CGAGGTGATCAACAAGCTGGGC); Rv2626c forward (CCGCGACATTGTGATCAAAG), reverse (GCTCTGAGATGACCGGAACAC), and probe (CGAACGCAAGCATCCAGGAGATGC); Rv1738 forward (CACTGGACCGTCGACATATCG), reverse (CGGTCGGCCGGATTG), and probe (CCAACGCAGCCGTGCCTTCG). Quantitative real-time PCR was performed on the LightCycler 480 (Roche). Residual DNA content was measured with reverse transcriptase negative controls and subtracted from each sample. All samples were normalized to the copy number of sigA present per cell. To determine copy number per cell, total RNA transcripts per sample were divided by the number of bacteria per ml of culture used to obtain the sample.
Mouse experiments
Six- to eight-week-old female specific-pathogen-free immunocompetent C57BL/6 mice (Charles River, Wilmington, MA) were inoculated with a low-dose (50–100 bacilli) aerosol of M. tuberculosis strains H37Rv, DorKO, or DorCO using a Glas-Col aerosol chamber. One day postinfection, three mice were sacrificed to verify bacterial uptake of 50 to 100 CFU per mouse. Following infection, mice were randomly divided into groups of 5–6 mice each. After the initial four weeks of infection, the untreated control groups were not treated with antibacterials in order to establish a chronic infection, and parallel groups were treated with the antibacterials INH and RIF administered in drinking water (at 100 and 60 mg/ml respectively) in order to establish a Cornell-type model of infection[33, 34] resulting in approximate in daily delivery of 25 mg/kg INH and 15 mg/kg RIF. Antibacterial therapy was continued for 12 weeks. Four weeks after cessation of treatment, immune suppression was initiated by administration of dexamethasone (6 mg/kg i.p. in 1×PBS, administered every two days) for another four weeks. Earlier time points consisted of five mice each, whereas later time points consisted of groups of 8–10 mice. Mice were sacrificed by CO2 inhalation. Spleens and left lungs were aseptically removed and disrupted in a tissue homogenizer. The number of viable organisms was determined by serial dilution of the homogenates on nutrient Middlebrook 7H11 agar plates (GIBCO BRL, Gaithersburg, MD). Plates were incubated at 37°C in ambient air for 4 weeks prior to the counting of viable M. tuberculosis colonies (CFU). After long-term treatment, the entire volume of each organ homogenate was plated to determine the total number of culturable mycobacteria per organ. For statistical analysis the viable counts were converted to logarithms, which were then evaluated by a one-way ANOVA followed by a multiple comparison analysis of variance by a one-way Tukey test (SigmaStat software program). Differences were considered significant at the 95% level of confidence.
Mouse lung histology
After euthanasia, caudal right lung lobes were infused in situ with 5 ml of 10% neutral-buffered formalin and preserved until processed for histopathological assessment. At the time of processing, all tissues were embedded in paraffin, sectioned at 5 µm, and stained with hematoxylin and eosin (H&E) for histologic evaluation and photography.
In vitro drug treatments
Logarithmic phase M. tuberculosis H37Rv growing at a starting OD600 of 0.1 were treated with either water, INH (1 µg/ml final concentration), RIF (0.5 µg/ml final concentration), or metronidazole (MET) (100 µg/ml final concentration) to confirm the effect of antibacterials on replicating bacilli. Cultures were sampled for viability at 48 hours post addition of antibiotics and plated in quadruplicate onto DTA agar plates and allowed to grow at 37°C for 2–3 weeks until colony formation was evident. Anaerobic dormant cultures were prepared as described above. Dormant cultures of H37Rv, DorKO, or DorCO were injected with water, INH (1 µg/ml final concentration), RIF (0.5 µg/ml final concentration), or MET (100 µg/ml final concentration) at day 4, 12, or 20 using a fixed needle insulin syringe. Cultures were then sampled 5 days after the addition of antibiotics, and were plated as described above to determine CFU counts.
Determination of DosR regulon orthologs
M. tuberculosis DosR regulon protein sequences were used to search a selection of bacterial genomes from the NCBI (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi) for candidate homologs, and the score of the top hit identified. Represented taxa included M. avium, M. paratuberculosis, M. leprae, M. bovis, M. tuberculosis, M. ulcerans, M. smegmatis, M. gilvum, M. vanbaalenii, Mycobacterium sp. KMS and Mycobacterium sp. JLS, Escherichia coli and Bacillus subtilis. The ORF identified by top hit for each species was then used as the query for blastp against the M. tuberculosis proteome; if blastp identified the initial M. tuberculosis gene this “reciprocal blast pair” was classified as a probable ortholog pair. Searches were automated using the Current Comparative Table (CCT) software [35].
Results and Discussion
Confirmation of DorKO and DorCO gene expression
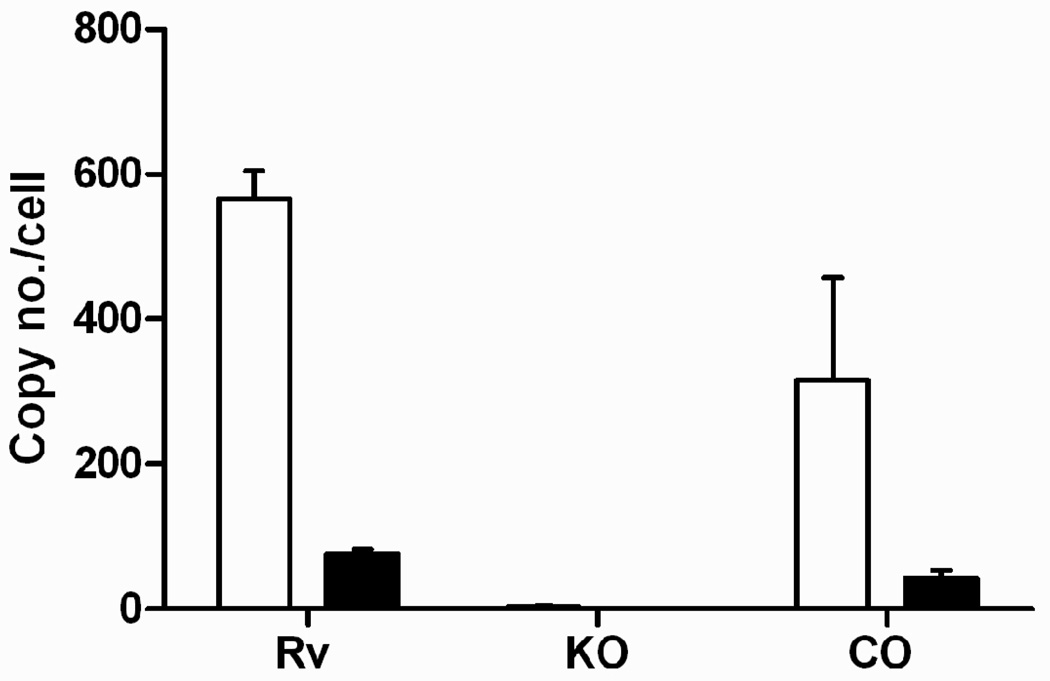
In order to study the role of the DosR regulon in antibacterial tolerance both in vitro and within a murine host, we first deleted the three-gene operon containing Rv3134c to Rv3132c, which includes dosR and two other DosR-controlled genes. Deletion of the entire three-gene operon made it possible during complementation to preserve all native regulatory elements, including internal promoters and operator sites that could be located within coding regions of adjacent genes. The absence of DosR regulon induction in the DorKO mutant and proper complementation in the DorCO strain were confirmed via microarray expression profiling and by growth and survival assays during anaerobic dormancy (data not shown) and qrt-PCR as reported here (Fig. 1). Wild type, DorKO and DorCO cultures were exposed to NO and mRNA was quantified by qrt-PCR. Gene induction of two representative DosR-controlled genes (Rv1738 and Rv2626) was confirmed in wild type and DorCO while gene induction was not observed in DorKO (Fig. 1).
Figure 1.
The DorKO mutant (KO) failed to induce two genes of the DosR regulon in response to nitric oxide as measured by qrt-PCR compared to H37Rv (Rv) and the DorCO (CO) strains. White bars indicate induction of Rv2626c and black bars induction of Rv1738. sigA transcripts were used to normalize RNA levels between samples.
Role of DosR regulon in chronic mouse infection
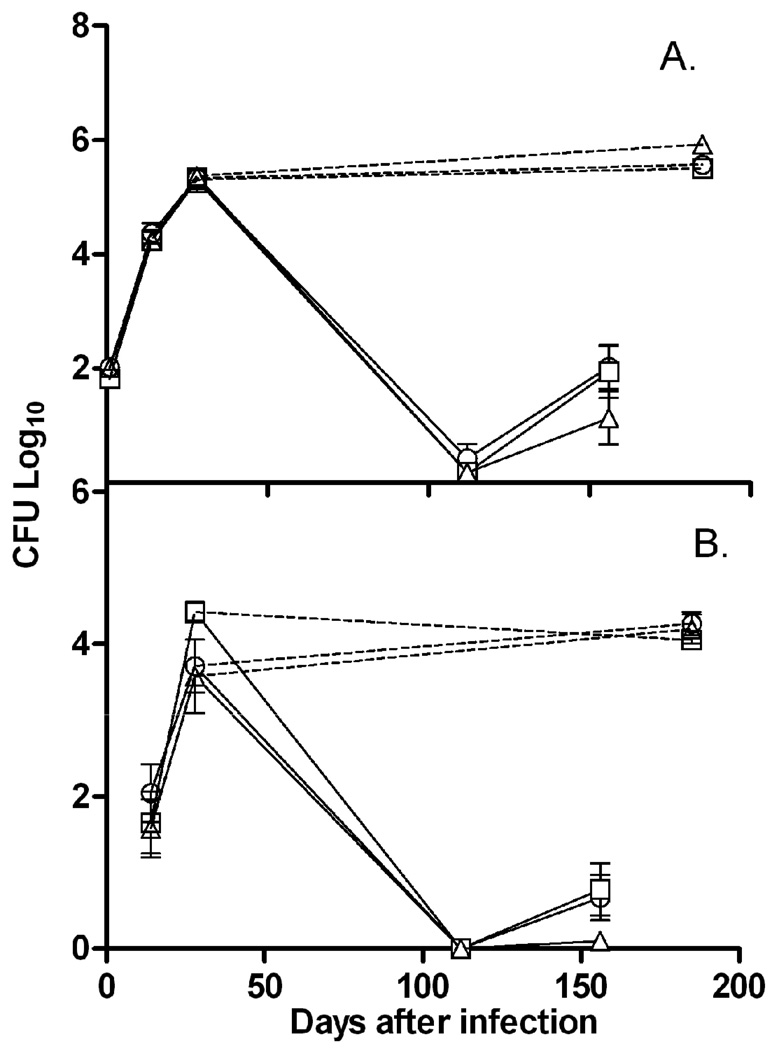
To determine the role of the DosR regulon in a chronic infection, C57BL/6 mice were infected with either wild-type H37Rv, the DorKO, or the DorCO by low dose aerosol infection of 50–100 bacilli. Mice were sacrificed at 24 hours, 2, 4 and 26 weeks. CFU counts from lungs (Fig. 2A) and spleens (Fig. 2B) were determined. As has been previously reported [36, 37], the regulon was not necessary for survival during long-term chronic mouse infection in either the lung or spleen. Interestingly, early in the spleen infection the DorKO mutant reached a bacterial burden of nearly 10-fold higher than that observed with the wild type and complemented strains; however, in the lungs the bacilli burden appears to be identical for all three strains (Fig. 2). After 6 months of infection the bacterial burden was nearly identical for all three strains in both the lungs and spleen. Parish et al. showed that a devR (dosR) disruption mutant grew to higher levels in the spleen, lungs, and liver of immunocompetent DBA mice early in infection, but reached similar levels in the lungs and spleens by day 60 [36]. Thus, our data partially support the earlier finding that a DosR mutant may be somewhat hypervirulent during initial stages of infection. However, we did not observe a higher bacterial load in DorKO within the lungs of the mice at any point during infection. The reason for the incongruity between these two studies is not clear, but could be due to variation in the mouse or bacterial strain used. Additionally, the differing route of infection used in the two studies could potentially explain the disparity between the results of the two studies, as the previous study used tail-vein, and our study used aerosol infection. A third study that also infected mice via aerosol found no difference in survival of a DosR mutant [37].
Figure 2.
Survival of the DorKO mutant during murine infection. C57BL/6 mice were inoculated with a low-dose (50–100 bacilli) aerosol of M. tuberculosis strains H37Rv (circles), DorKO (squares), or DorCO (triangles). The chronic (dashed lines) and Cornell model (solid lines) groups ran parallel for the initial 4 weeks of infection, at which point mice of the Cornell model groups were treated with the antibacterials INH and RIF (administered in drinking water). After 12 weeks, antimicrobial therapy was discontinued and four weeks after cessation of treatment, immune suppression was initiated by administration of dexamethasone for four weeks. CFU counts were determined for (A) the lung or (B) the spleen. For the earlier time points every group consisted of five mice, while for later time points groups consisted of 8–10 mice.
Although the DosR regulon is highly expressed in mice [9, 22], DosR does not play a survival role in this in vivo environment. This result is not surprising in view of the fact that the mouse lung is not hypoxic, and the mouse model itself, although relatively inexpensive and convenient, is not necessarily a faithful representation of the course of infection within the human lung [38–41]. Recent studies have also demonstrated that bacilli continue to replicate and maintain metabolic activity in murine infections [42]. These findings indicate that bacilli either maintain a high level of replication during latent infection or the chronic murine model fails to capture critical elements of latent infection. Although mice form granulomas when infected with M. tuberculosis, the pathology of these granulomas is unlike that seen in human infection [38, 43]. Interestingly, Malhotra et al., observed that a dosR mutant was attenuated in the guinea pig model [44]. The Guinea pig model more closely represents human pathology and produces anoxic lesions [41, 45–47]. A recent study by Converse et al. also demonstrated a DosR mutant was defective in Guinea pig and rabbit infections [48]. These results suggest that the regulon may be important for in vivo survival during conditions where lesions become oxygen restricted.
Reduced lung pathology in DosR regulon mutant
The pathology was quite comparable between the three strains when analyzed after two and four weeks post aerosol infection (data not shown). However, mouse lungs infected for six months with H37Rv, DorKO or DorCO showed that lymphocyte infiltration and granuloma formation was hampered in mice infected with DorKO (Supplemental Fig. 1). The number of granulomas was almost 50% less versus H37Rv, and the pathological severity was also dramatically reduced. Lymphocytes in lungs infected with the DorKO mutant reside in peribronchial and perivascular regions and, apart from one animal, have not infiltrated deeper into the lungs as is evident in lungs infected with wild type or DorCO strains. DorKO infected lungs also show less thickening of the parenchyma and the presence of foamy macrophages. For the DorCO strain, pathology was slightly less severe compared to the wild type H37Rv. The overall cellular composition of the granulomas induced by all three strains showed a similar accumulation of lymphocytes and macrophages. These data complement two previous studies that demonstrate less pathology for a DosR regulon mutant compared to wild type. Malhotra et al. first observed this phenotype in Guinea pig infections and Converse et al. recently observed the same phenomenon in murine, Guinea pig, and rabbit infections [44, 48]. These findings indicate the DosR regulon has effects on the immune system but the mechanism is unknown.
Role of DosR regulon in drug tolerance during mouse infection
To deduce the importance of the DosR regulon to antibacterial tolerance in the context of a host infection, we employed the Cornell model which attempts to mimic a latent infection in humans by eliminating growing bacilli with antibacterial treatment [33, 34]. C57BL/6 mice were inoculated via aerosol, and INH and RIF therapy was started four weeks after infection to reduce the bacterial numbers below the detection limit and eliminate replicating bacilli. Following 12 weeks of drug therapy, treatment was discontinued for four weeks (as was indicated by McCune et al. as being necessary for relapse), after which dexamethasone was administered to suppress immunity and allow for rapid growth of any remaining viable bacilli. As shown in Figure 2, the survival of H37Rv and mutant DorKO bacilli were indistinguishable. In addition, it appeared that DorCO did less well in reactivating after drug treatment in both the lungs and spleen (Fig. 2). These data indicate that DosR is not required for M. tuberculosis tolerance to antibacterial therapy during a mouse infection as a subpopulation is able to withstand 12 weeks of INH and RIF treatment even in the absence of regulon induction.
Role of the DosR regulon in drug tolerance during anaerobic dormancy
Since the mouse model of M. tuberculosis infection is not an anoxic environment and does not fully mimic a human infection[38–40] and does not appear to require the DosR regulon, we sought to measure antibacterial indifference under conditions known to induce and require the DosR regulon and which are drug tolerant. To this aim we used the “Wayne model” of anaerobic dormancy in which bacilli adapted to anaerobic conditions strongly induce DosR and concomitantly become indifferent to RIF and INH[15, 26]. When anaerobic dormancy is performed as described by Wayne and Hayes [15], significant death is not observed in our DosR mutant until later than 20 days in the model. However, we observed rapid death of the DorKO mutant when a model similar to that described by Boon and Dick was used [19]. This model incorporates fast stirring and results in nearly a 10,000-fold defect in the DorKO mutant by day 40 (data not shown). In this study we used the original Wayne model as it was the model used in previous studies that demonstrated drug tolerance of anaerobic M. tuberculosis. The DosR regulon is also known to be highly expressed in this model [10], and early death does not occur in the DorKO mutant that could obscure the effects of the drugs.
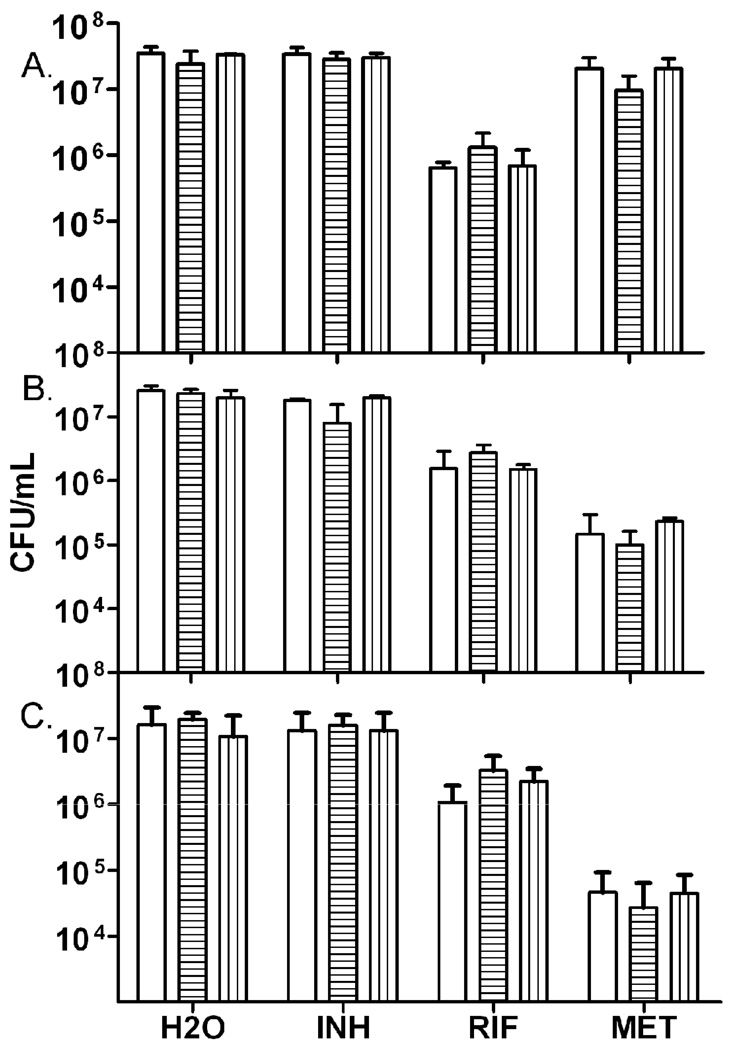
Dormancy model tubes containing M. tuberculosis were injected with either water, INH, RIF, or MET at 4, 12, and 20 days into the model. In this model bacterial respiration consumed the available oxygen in the sealed tubes in approximately 12 days, at which point oxygen levels dropped below the concentration necessary to support bacilli growth [15]. Samples were removed five days post treatment to determine bacilli survival. As shown in Figure 3, the DorKO mutant was no more susceptible to these antibacterials than H37Rv or the complemented strain at any time point during the dormancy model. Previous reports [15, 26] were used to guide the selection of antibiotics and conditions used in this work, and as in those studies, wild type H37Rv cultures grown aerobically were susceptible to INH and RIF but not MET (data not shown). Although the DosR regulon is required for long-term survival of the bacilli during a dormant state [14, 19], it is not directly required for drug tolerance observed in this study.
Figure 3.
Survival of H37Rv, the DorKO, or the DorCO during the Wayne model of dormancy after antibacterial challenge. H37Rv (white bars), DorKO (horizontal stripes), or DorCO (vertical stripes) were challenged with H2O, Isoniazid (INH), Rifampin (RIF), or Metronidazole (MET) on (a) day four, (b) day 12, or (c) day 20 into the model. CFU were determined by plating onto DTA plates five days post addition of drug.
Presence of DosR regulon orthologs in Mycobacteriaceae
To obtain a better understanding of the role for the DosR regulon we searched related bacterial species for candidate homologs and/or orthologs. The existence of regulon orthologs was determined in other mycobacteria as well as in the model Gram-positive and -negative bacteria B. subtilis and E. coli (Fig. 4). As expected, most regulon proteins are highly conserved in M. bovis and many are in M. ulcerans, which is a pathogen but also exists in an environmental reservoir. There is also broad conservation of regulon genes across the rapid-growing environmental mycobacteria. By contrast there is much less conservation of DosR-regulated proteins in three non-tuberculosis pathogens; M. avium, M. paratuberculosis, and M. leprae. Conservation in M. leprae is in the range of those found in non-mycobacteria species such as B. subtilis and E. coli.
Figure 4.
A graphical representation of DosR regulon conservation in Mycobacteriaceae and beyond. Rows represent M. tuberculosis ORFs and columns represent sample taxa, organized by phylogenetic relationship, where known[51]. Represented taxa included M. avium (A), M. paratuberculosis (P), M. leprae (L), M. bovis (B), M. tuberculosis (T), M. ulcerans (U), M. smegmatis (S), M. gilvum (G), M. vanbaalenii (V), Mycobacterium sp. KMS (K), M. marinum (M), Mycobacterium sp. JLS (J), Escherichia coli (Ec) and Bacillus subtilis (Bs). Homologs of M. tuberculosis genes were detected using tblastn to search each strain’s genome. Genomes containing a top hit greater than e−5 are shaded grey, and greater than e−10 are striped to indicate the presence of one or more homologous sequences. The ORF identified by the top hit in each species was then used to blastp the M. tuberculosis proteome. If it identified the initial M. tuberculosis gene, this “reciprocal blast pair” was classified as a probable ortholog pair and the corresponding cell colored black.
Conclusions
DosR is necessary for the induction of a 48 gene regulon[10, 14], whose expression allows long term survival under anaerobic conditions, [14, 19]. Bacilli in an anaerobic state are also indifferent to several first line M. tuberculosis antibacterials[15, 26]. These facts have led to widespread suggestions that dormancy and perhaps the DosR regulon itself may contribute to the high level of M. tuberculosis drug tolerance during infection [2, 3, 5–7, 12, 15, 21, 28–30, 49, 50]. The data presented here demonstrate that the DosR regulon does not play a central role in antibacterial tolerance, at least not for tolerance to the specific drugs tested in murine infection or during early anaerobic dormancy. In the absence of induction of the DosR regulon, the survival of mutant and wild type bacilli are indistinguishable in murine models of drug treatment. If non-respiring dormancy is important in aspects of drug tolerance during human infection, the mouse model does not capture these aspects of the natural infection. It is possible that a subpopulation of bacilli exists in a dormant-like state during human infection and survives due to the DosR regulon. However, the fact that they are not replicating or metabolically dormant specifically allows them to tolerate antibacterials. Thus, without the regulon this population of bacilli would be eliminated and not contribute to a disease relapse after treatment.
The presence of much of the DosR regulon in environmental mycobacteria suggests that it did not evolve specifically for survival within the host or due to therapeutic antibiotic stress. Instead the pattern we observe is more consistent with the possibility that the regulon evolved to deal with conditions within the environment such as anaerobiosis. If true, once the ancestor of modern M. tuberculosis found itself inside a human host, the DosR gene products may have provided increased protection from the immune system or allowed the bacilli to survive in a specific niche in a growth arrested state. The fact that the highly expressed and thus costly DosR regulon has been conserved in M. tuberculosis, but not other pathogenic mycobacteria lacking a latent stage of infection, further suggests DosR regulon imparts important advantages tied to latency. Phylogenetic analysis of individual genes should provide a method of testing these and other hypotheses in the future.
The findings herein indicate that targeting the DosR regulon may be beneficial but would not be sufficient to overcome the intractable problem of drug tolerance.
Supplementary Material
Murine lung histopathology 6 months post-infection. Granuloma formation in lungs from M. tuberculosis infected mice sacrificed 6 months post infection. Magnification 20× and 100×, hematoxylin and eosin stained.
Acknowledgements
This research project was funded by NIH grant RO1 AI061505 entitled “Mycobacterium tuberculosis dormancy program” awarded to M.I.V. and A.L., NIH grant RO1 AI44826 awarded to G.K.S., NIH grant R15 AI068706 awarded to R.R., and T32 AI052066-07 awarded to I.L.B. Additional funding was provided by the American Lung Association Dalsemer Career Development Award awarded to M.I.V. We thank Hannah Franklin (Seattle University) for data analysis and the staff of the Laboratory Animal Resources (Colorado State University) for animal care.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dye C, et al. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. Jama. 1999;282(7):677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed]
- 2.Wallis RS, et al. Drug tolerance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1999;43(11):2600–2606. doi: 10.1128/aac.43.11.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallis RS, Palaci M, Eisenach K. Persistence, not resistance, is the cause of loss of isoniazid effect. J Infect Dis. 2007;195(12):1870–1871. doi: 10.1086/518044. author reply 1872-3. [DOI] [PubMed] [Google Scholar]
- 4.Mitchison DA, et al. Isoniazid activity is terminated by bacterial persistence. J Infect Dis. 2007;195(12):1871–1872. doi: 10.1086/518046. author reply 1872-3. [DOI] [PubMed] [Google Scholar]
- 5.Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb) 2004;84(1–2):29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Parrish NM, Dick JD, Bishai WR. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 1998;6(3):107–112. doi: 10.1016/s0966-842x(98)01216-5. [DOI] [PubMed] [Google Scholar]
- 7.Dick T. Dormant tubercle bacilli: the key to more effective TB chemotherapy? J Antimicrob Chemother. 2001;47(1):117–118. doi: 10.1093/jac/47.1.117. [DOI] [PubMed] [Google Scholar]
- 8.Leyten EM, et al. Human T-cell responses to 25 novel antigens encoded by genes of the dormancy regulon of Mycobacterium tuberculosis. Microbes Infect. 2006;8(8):2052–2060. doi: 10.1016/j.micinf.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Schnappinger D, et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med. 2003;198(5):693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voskuil MI. Mycobacterium tuberculosis gene expression during environmental conditions associated with latency. Tuberculosis (Edinb) 2004;84(3–4):138–143. doi: 10.1016/j.tube.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 2004;84(3–4):218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Wayne LG, Sohaskey CD. Nonreplicating persistence of mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 13.Bardarov S, et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148(Pt 10):3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 14.Park HD, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48(3):833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64(6):2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin I, Voskuil R, WHaAJCS . Oxygen, Nitric Oxide and Carbon Monoxide Signalling. In: Brown TPaA., editor. Mycobacterium: Genomics and Molecular Biology. London: Caister Academic Press; 2008. pp. 121–147. [Google Scholar]
- 17.Kendall SL, et al. The Mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis (Edinb) 2004;84(3–4):247–255. doi: 10.1016/j.tube.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Roberts DM, et al. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J Biol Chem. 2004;279(22):23082–23087. doi: 10.1074/jbc.M401230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boon C, Dick T. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J Bacteriol. 2002;184(24):6760–6767. doi: 10.1128/JB.184.24.6760-6767.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rachman H, et al. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun. 2006;74(2):1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warner DF, Mizrahi V. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin Microbiol Rev. 2006;19(3):558–570. doi: 10.1128/CMR.00060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roupie V, et al. Immunogenicity of eight dormancy regulon-encoded proteins of Mycobacterium tuberculosis in DNA-vaccinated and tuberculosis-infected mice. Infect Immun. 2007;75(2):941–949. doi: 10.1128/IAI.01137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell IA, Bah-Sow O. Pulmonary tuberculosis: diagnosis and treatment. Bmj. 2006;332(7551):1194–1197. doi: 10.1136/bmj.332.7551.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbert D, et al. Bactericidal action of ofloxacin, sulbactam-ampicillin, rifampin, and isoniazid on logarithmic- and stationary-phase cultures of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1996;40(10):2296–2299. doi: 10.1128/aac.40.10.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spigelman MK. New tuberculosis therapeutics: a growing pipeline. J Infect Dis. 2007;196 Suppl 1:S28–S34. doi: 10.1086/518663. [DOI] [PubMed] [Google Scholar]
- 26.Wayne LG, Sramek HA. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38(9):2054–2058. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munoz-Elias EJ, et al. Replication dynamics of Mycobacterium tuberculosis in chronically infected mice. Infect Immun. 2005;73(1):546–551. doi: 10.1128/IAI.73.1.546-551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy DJ, Brown JR. Novel drug target strategies against Mycobacterium tuberculosis. Curr Opin Microbiol. 2008 doi: 10.1016/j.mib.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Connolly LE, Edelstein PH, Ramakrishnan L. Why is long-term therapy required to cure tuberculosis? PLoS Med. 2007;4(3):e120. doi: 10.1371/journal.pmed.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed MB, et al. The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J Bacteriol. 2007;189(7):2583–2589. doi: 10.1128/JB.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stover CK, et al. New use of BCG for recombinant vaccines. Nature. 1991;351(6326):456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 32.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198(5):705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCune RM, et al. Microbial persistence. I. The capacity of tubercle bacilli to survive sterilization in mouse tissues. J Exp Med. 1966;123(3):445–468. doi: 10.1084/jem.123.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCune RM, Jr., McDermott W, Tompsett R. The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by the administration of pyrazinamide and a companion drug. J Exp Med. 1956;104(5):763–802. doi: 10.1084/jem.104.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landsteiner BR, Olson MR, Rutherford R. Current Comparative Table (CCT) automates customized searches of dynamic biological databases. Nucleic Acids Res. 2005;33(Web Server issue):W770–W773. doi: 10.1093/nar/gki432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parish T, et al. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect Immun. 2003;71(3):1134–1140. doi: 10.1128/IAI.71.3.1134-1140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rustad TR, et al. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS ONE. 2008;3(1):e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter RL, Jagannath C, Actor JK. Pathology of postprimary tuberculosis in humans and mice: contradiction of long-held beliefs. Tuberculosis (Edinb) 2007;87(4):267–278. doi: 10.1016/j.tube.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Orme IM. The mouse as a useful model of tuberculosis. Tuberculosis (Edinb) 2003;83(1–3):112–115. doi: 10.1016/s1472-9792(02)00069-0. [DOI] [PubMed] [Google Scholar]
- 40.Aly S, et al. Oxygen status of lung granulomas in Mycobacterium tuberculosisinfected mice. J Pathol. 2006;210(3):298–305. doi: 10.1002/path.2055. [DOI] [PubMed] [Google Scholar]
- 41.Flynn JL. Lessons from experimental Mycobacterium tuberculosis infections. Microbes Infect. 2006;8(4):1179–1188. doi: 10.1016/j.micinf.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 42.Gill WP, et al. A replication clock for Mycobacterium tuberculosis. Nat Med. 2009;15(2):211–214. doi: 10.1038/nm.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhoades ER, Frank AA, Orme IM. Progression of chronic pulmonary tuberculosis in mice aerogenically infected with virulent Mycobacterium tuberculosis. Tuber Lung Dis. 1997;78(1):57–66. doi: 10.1016/s0962-8479(97)90016-2. [DOI] [PubMed] [Google Scholar]
- 44.Malhotra V, et al. Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol Lett. 2004;231(2):237–245. doi: 10.1016/S0378-1097(04)00002-3. [DOI] [PubMed] [Google Scholar]
- 45.Turner OC, Basaraba RJ, Orme IM. Immunopathogenesis of pulmonary granulomas in the guinea pig after infection with Mycobacterium tuberculosis. Infect Immun. 2003;71(2):864–871. doi: 10.1128/IAI.71.2.864-871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Via LE, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76(6):2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenaerts AJ. Location of persisting mycobacteria in a Guinea pig model of tuberculosis revealed by r207910. Antimicrob Agents Chemother. 2007;51(9):3338–3345. doi: 10.1128/AAC.00276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Converse PJ. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect Immun. 2009;77(3):1230–1237. doi: 10.1128/IAI.01117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bloom BR, McKinney JD. The death and resurrection of tuberculosis. Nat Med. 1999;5(8):872–874. doi: 10.1038/11309. [DOI] [PubMed] [Google Scholar]
- 50.Stewart GR, Robertson BD, Young DB. Tuberculosis: a problem with persistence. Nat Rev Microbiol. 2003;1(2):97–105. doi: 10.1038/nrmicro749. [DOI] [PubMed] [Google Scholar]
- 51.Tortoli E. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin Microbiol Rev. 2003;16(2):319–354. doi: 10.1128/CMR.16.2.319-354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Murine lung histopathology 6 months post-infection. Granuloma formation in lungs from M. tuberculosis infected mice sacrificed 6 months post infection. Magnification 20× and 100×, hematoxylin and eosin stained.