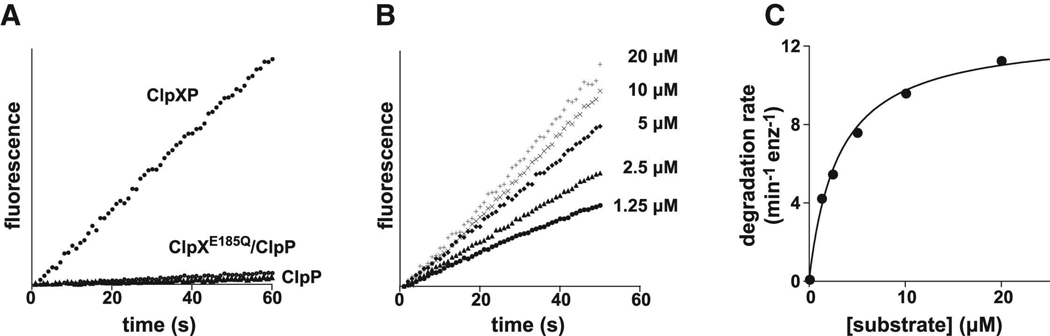
Figure 2.
(A) Efficient cleavage of the [G10] peptide by ClpP was observed in the presence of wild-type ClpX but not in the absence of ClpX or with ClpXE185Q, which cannot hydrolyze ATP. All reactions contained 10 µM of the [G10] substrate and 300 nM ClpP14. When present, the concentration of ClpX6 or the ATPase-defective mutant was 800 nM. (B) Degradation of different concentrations of the [VG]5 peptide by 800 nM ClpX6 and 300 nM ClpP14. (C) Steady-state rates of [VG]5 peptide degradation by ClpXP were calculated from the data in panel C and fit to the Michaelis-Menten equation (KM = 3.1 µM; Vmax = 12.7 min−1 ClpP−1).