Fig. 3.
The rpt4-G106D and nas2Δ mutants have proteasome assembly defects.
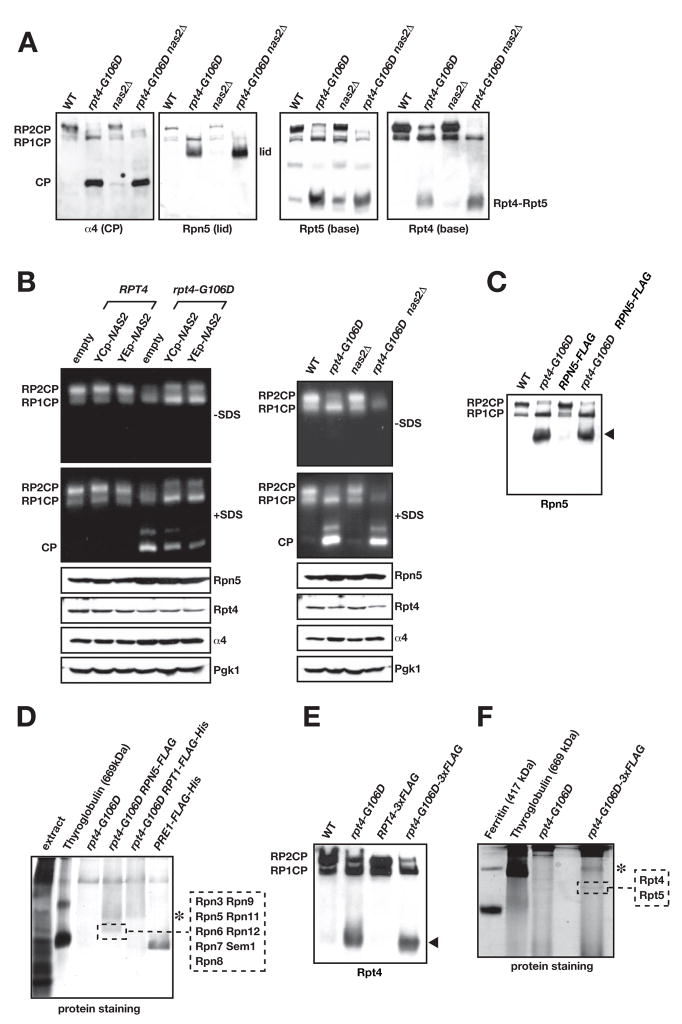
A. Immunoblot analysis of nondenaturing PAGE-separated proteins from yeast whole-cell extracts. The faint central band in the anti-Rpt5 blot is of unknown composition.
B. Nondenaturing gel separation of proteins from yeast extracts followed by gel overlay with the fluorogenic substrate Suc-LLVY-AMC to assay proteasomal activity. Left panel, effect of low-copy (YCp) and high-copy (YEp) expression of NAS2 on functional proteasome formation. Right panel, effect of nas2Δ on assembly. Addition of SDS to the reaction buffer activates the otherwise latent CP and Blm10-CP (unlabeled band above free CP).
C. A C-terminal Flag tag on Rpn5 does not disrupt the Rpn5-containing RP subcomplex in rpt4-G106D cells (arrowhead).
D. Nondenaturing gel separation of Flag-affinity purified proteins from rpt4-G106D RPN5-FLAG cells. MS/MS analysis of the indicated band from the GelCode Blue-stained gel demonstrated that it was the 9-subunit RP lid. Asterisk, a band that is likely the free RP but was not analyzed. Unlike panel C, gel separation included a stacking gel and was run longer.
E. A triplicated Flag tag on Rpt4 does not disrupt the Rpt4-containing RP subcomplex in rpt4-G106D cells (arrowhead).
F. Nondenaturing gel separation of Flag-affinity purified proteins from rpt4-G106D RPT4-3xFLAG cells. MS/MS analysis of the indicated band from the GelCode Blue-stained gel yielded Rpt4 and Rpt5 from the RP base. Asterisk, as in panel D. PAGE was done with a 5.5% PA separating gel instead of the 4% used in other panels, and a stacking gel was used.