Fig. 1. Pathways of arsenic and selenium uptake and efflux in prokaryotes and eukaryotes.
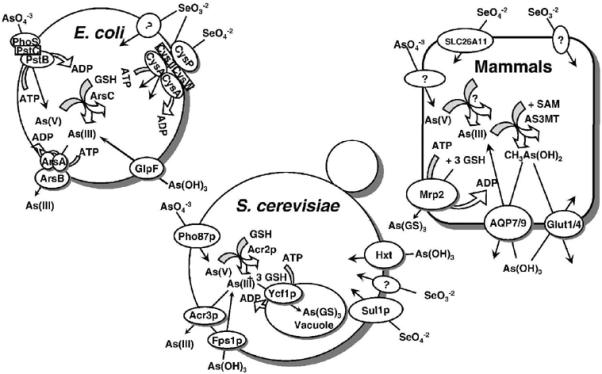
Arsenate (As(V)) is taken up by phosphate transporters, while As(III) is taken up by aquaglyceroporins (GlpF in E. coli, Fps1p in yeast, and AQP7 and AQP9 in mammals), and hexose permeases (HXT1, HXT3, HXT4, HXT5, HXT7, or HXT9 in yeast, and GLUT1 and GLUT4 in mammals). In both E. coli and S. cerevisiae, arsenate is reduced to arsenite by the bacterial ArsC or yeast Acr2p enzymes. In both organisms, glutathione and glutaredoxin serve as the source of reducing potential. The proteins responsible for arsenate uptake and reduction in mammals have not yet been identified. In E. coli, arsenite is extruded from the cells by ArsB alone or by the ArsAB ATPase. In yeast Acr3p is a plasma membrane arsenite efflux protein, and Ycf1p, which is a member of the MRP family of the ABC superfamily of drug-resistance pumps, transports As(GS)3 into the vacuole. In mammals, Mrp isoforms such as Mrp2, pump As(GS)3 out of cells. Selenate is taken up by sulfate permeases, the CysAWTP ABC ATPase in bacteria, Sul1p in yeast and SLC26A11 in humans. By-and-large, the uptake pathways for selenite have not been identified.
