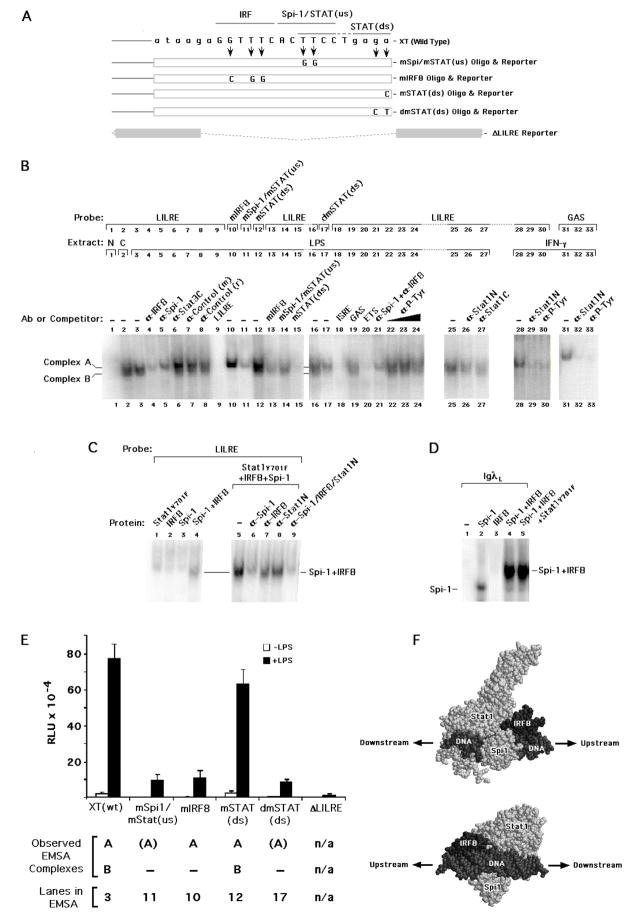
Fig. 3.
Functional and proposed structural nature of the LILRE. (A) Schematic showing the location of the individual mutations in binding sites within a long construct harboring a 3.76 kb region of the human IL1B gene, designated as XT. Similar mutations were made in the labeled probes and cold competitors used for EMSA. These sites include an IRF (mIRF) site, the Spi-1 site (which overlaps the upstream STAT/GAS half-site) designated as mSpi-1/mSTAT(us), and both single and double point mutations designated as mSTAT(ds) and dmSTAT(ds), respectively, in the downstream STAT half-site. An XT-derived construct in which the entire LILRE sequence was deleted is denoted as Δ LILRE. (B) Images of relevant EMSA data showing the binding of complexes from nuclear extracts derived from both control cells (labeled C) and cells exposed for 30 min to LPS or IFNγ. Binding studies used wild type and mutated (as indicated) LILRE or GAS α-32P-labeled probes at a final ionic strength of 50 mM Na+ plus K+. Null binding reactions (labeled N) did not contain extract. Antibodies were pre-incubated with nuclear extract on ice prior to the addition of labeled probe. Unlabeled competitor oligonucleotides were used at a 100-fold molar excess over the molar concentration of the probe. (C) EMSA showing the binding of in vitro expressed proteins to wild type LILRE α-32P-labeled probes at an ionic strength of 50 mM Na+, optimal for Spi-1 binding(Galson and Housman, 1988). Antibodies were added as indicated before addition of probe. (D) EMSA showing the binding of in vitro expressed proteins to IgλL α-32P-labeled probe. (E) Activity of wild type and mutant IL1B gene reporter in RAW 264.7 cells transfected with wild type XT, mIRF XT, mSpi-1/STAT(us) XT, mSTAT(ds) XT, dmSTAT(ds) XT, and ΔLILRE XT, each at 250 ng with total DNA adjusted to 300 ng with empty pGL3Basic vector. Cells were transfected with the indicated constructs and either left untreated or treated with 10 μg/ml LPS for 4 hrs. The values are derived from the average of three independent experiments. (F) Hypothetical arrangement of IRF8, Spi-1 and non-tyrosine phosphorylated (NTP) Stat1 bound to LILRE DNA. Proteins and DNA are labeled and represented as space filling models. The orientation of the proteins is indicated by arrows pointing upstream and downstream, either toward or away from, respectively, the transcription start site. The lower image is related to the upper via a 180° rotation about the Y axis plus a −70° rotation about the X axis.