Abstract
Wall-bound purple acid phosphatases have been shown to be potentially involved in the regulation of plant cell growth. The aim of this work was to further investigate the function of one of these phosphatases in tobacco (Nicotiana tabacum), NtPAP12, using transgenic cells overexpressing the enzyme. The transgenic cells exhibited a higher level of phosphatase activity in their walls. The corresponding protoplasts regenerating a cell wall exhibited a higher rate of β-glucan synthesis and cellulose deposition was increased in the walls of the transgenic cells. A higher level of plasma membrane glucan synthase activities was also measured in detergent extracts of membrane fractions from the transgenic line, while no activation of Golgi-bound glycan synthases was detected. Enzymatic hydrolysis and methylation analysis were performed on the products synthesized in vitro by the plasma membrane enzymes from the wild-type and transgenic lines extracted with digitonin and incubated with radioactive UDP-glucose. The data showed that the glucans consisted of callose and cellulose and that the amount of each glucan synthesized by the enzyme preparation from the transgenic cells was significantly higher than in the case of the wild-type cells. The demonstration that callose and cellulose synthases are activated in cells overexpressing the wall-bound phosphatase NtPAP12 suggests a regulation of these carbohydrate synthases by a phosphorylation/dephosphorylation process, as well as a role of wall-bound phosphatases in the regulation of cell wall biosynthesis.
Plant cell walls contain acid phosphatases (EC 3.1.3.2) that are potentially involved in regulating cell growth processes. Recently, we suggested that the wall-bound purple acid phosphatase NtPAP12 from tobacco (Nicotiana tabacum) functions as a protein phosphatase (Kaida et al., 2008) and promotes cellulose synthesis (Sano et al., 2003). Its transcription level was significantly increased in protoplasts during wall regeneration (Kaida et al., 2003). Interestingly, addition of the phosphatase to the protoplasts accelerated β-glucan deposition, whereas the addition of antibodies directed against the phosphatase prevented β-glucan deposition (Sano et al., 2003). Part of the β-glucans formed by tobacco protoplasts during the early stage of wall regeneration consist of β-1,4-glucans (Shea et al., 1989). It was hypothesized that the same enzyme may be involved in the biosynthesis of callose and cellulose and that the biosynthesis of either polysaccharide may be controlled by phosphorylation (Delmer, 1999). However, experimental data showed that the cellulose and callose synthase catalytic subunits are encoded by different genes (Pear et al., 1996; Arioli et al., 1998; Cui et al., 2001; Doblin et al., 2001; Hong et al., 2001a; Li et al., 2003; Brownfield et al., 2007). Indeed, these proteins show no significant sequence similarity and are grouped in different glycosyltransferase families in the CAZY database (http://www.cazy.org), family 2 for cellulose synthase and family 48 for callose synthase (Cantarel et al., 2009). Thus, it is currently admitted that callose and cellulose synthases correspond to two different enzyme proteins. Notwithstanding the existence of two different enzymes catalyzing the polymerization of callose and cellulose, it is possible that the two activities are coregulated by phosphorylation/dephosphorylation, but this remains to be demonstrated. Several putative cytoplasmic phosphorylation sites have been identified in cellulose synthase in Arabidopsis (Arabidopsis thaliana) using a phosphoproteomics approach (Nühse et al., 2004) but their regulatory function was not determined. A more recent report on the identification of the phosphorylation sites of AtCesA7, which is a cellulose synthase subunit involved in secondary cell wall formation in Arabidopsis, points toward a role of phosphorylation events in regulating the turnover of cellulose synthase by proteolysis through a proteasome-dependent pathway (Taylor, 2007). The implication of phosphorylation in the regulation of β-glucan synthases has also been proposed in pea (Pisum sativum) stems (Ray, 1973). In addition, sodium fluoride, which inhibits the action of phosphoprotein phosphatases was shown to increase by 5-fold β-glucan synthase activity from corn (Zea mays) in the presence of Ca2+, possibly in a calmodulin-dependent manner (Paliyath and Poovaiah, 1988). Overall, phosphorylation seems to play an important role in the regulation of the activities of plant β-glucan synthases, including cellulose synthase, but the underlying molecular mechanisms and resulting effects are poorly understood.
To shed light on the possible function of NtPAP12 in the regulation of β-glucan and cellulose biosynthesis, expression of the phosphatase was induced at the early stage of wall regeneration in tobacco protoplasts (Kaida et al., 2003). Here we have exploited this system to demonstrate both in vivo and in vitro that overexpression of NtPAP12 activates the plasma membrane-bound cellulose and callose synthases, while Golgi β-glucan synthases and xyloglucan xylosyltransferases were not affected. Our work reveals that NtPAP12 is involved in the regulation of callose and cellulose synthases and, indirectly, in cell growth and morphogenesis.
RESULTS
Localization of Purple Acid Phosphatase and β-Glucans in Tobacco Protoplasts during Wall Regeneration
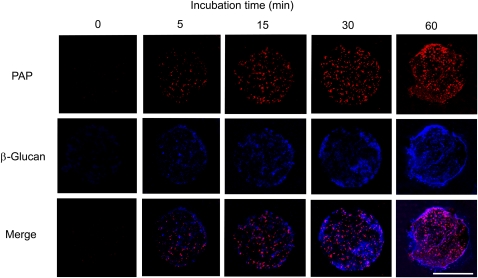
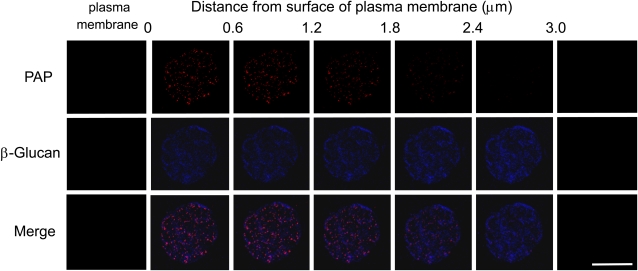
To determine the localization of NtPAP12 in protoplasts undergoing cell wall regeneration, an aliquot of the protoplast suspension overexpressing the phosphatase gene was transferred to a coverslip coated with polylysine and incubated in the medium for wall regeneration. At the early stages of regeneration (0–60 min), the cells were subjected to lysis with a low osmotic buffer to obtain immobilized disc-shaped protoplast ghosts designated as plasma membrane sheets (Hirai et al., 1998). A double detection based on immunofluorescence labeling of purple acid phosphatase and Calcofluor White staining of β-glucans was subsequently performed. Fluorescence microscopy showed that the purple acid phosphatase could be visualized between the plasma membrane sheets and the polylysine-coated coverslip immediately after the beginning of the incubation, and its deposition increased with incubation time (Fig. 1). β-Glucan microstructures had also been formed on the plasma membrane sheets between the sheets and the polylysine-coated coverslip (Fig. 1). The β-glucan stained with Calcofluor White first emerged as dots at many loci within a few minutes after the beginning of the incubation, and short microstructures, most likely microfibril aggregates, were clearly visible after about 15 min of incubation. Merged images showed that phosphatase deposition and β-glucan formation seem to occur near the surface of the plasma membrane (Fig. 1). A series of optical section images were recorded using steps of 0.6 μm depth from the membrane to the outside of the walls (Fig. 2). Purple acid phosphatase was detected in the walls with a higher intensity in the sections closest to the membrane. The intensity of the signal gradually decreased in the outer layers of the scan. It seems likely that the phosphatase is essentially present in close proximity to the plasma membrane (Fig. 2). The β-glucan microstructures appeared to be uniformly distributed at the surface of the membrane and formed a 3-μm thick extracellular layer (Fig. 2).
Figure 1.
Deposition of purple acid phosphatase (PAP) and β-glucan on plasma membrane sheets during time course. Plasma membrane sheets were prepared from protoplasts cultured in the wall-regeneration medium for 0, 5, 15, 30, and 60 min. Purple acid phosphatase was detected with anti-wall-bound purple acid phosphate antibodies and β-glucan was stained with Calcofluor White. Each image was joined from five 0.6 μm optical section images. Bar = 50 μm.
Figure 2.
Series of optical section images (0.6 μm) from the plasma membrane surface to the outer part of the cell wall. Plasma membrane sheets were prepared from protoplasts cultured in the wall-regeneration medium for 30 min. Purple acid phosphatase (PAP) was detected using anti-wall-bound purple acid phosphate antibody and β-glucan was stained with Calcofluor. Bar = 50 μm.
Levels of Expression of Phosphatase and β-Glucan Deposition at the Surface of the Protoplasts
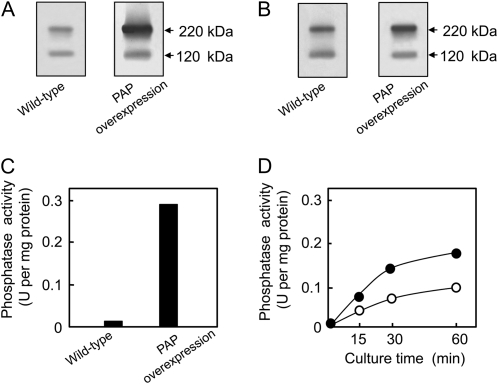
We produced transgenic tobacco cells (XD-6) that overexpressed NtPAP12 under the control of an enhanced constitutive promoter, the cauliflower mosaic virus 35S promoter (Mitsuhara et al., 1996), to study the role of purple acid phosphatase in the walls of tobacco cells. Transgenic cells overexpressing NtPAP12 and wild-type cells at logarithmic phase were harvested and the levels of expression of the enzyme were measured by both western blot using antiphosphatase antibodies and assaying phosphatase activity. Considerably more of the 220-kD polypeptide was present in the walls of the transgenic cells than in those of the wild-type cells (Fig. 3A). Although the amount of the 120-kD polypeptide in the walls of the transgenic and wild-type cells was comparable, as judged by western blotting, the amount of the 220-kD polypeptide was significantly higher in the walls of the transgenic cells compared to those of the wild-type cells. The phosphatase occurs as a 120-kD dimer composed of two subunits of 60 kD in the Golgi, and this dimer is translocated to the walls where it forms a tetramer of 220 kD (Kaida et al., 2008). The activity of the phosphatase was 20-fold higher in the walls of the transgenic cells compared to the wild-type cells (Fig. 3C). Consistent with the induction of NtPAP12 during wall regeneration (Kaida et al., 2003), the level of the 220-kD polypeptide was rather high in the regenerating walls of the wild-type cells and even higher in those of the transgenic cells (Fig. 3). The levels of activity in the nascent walls on the protoplasts overexpressing NtPAP12 doubled at the early stage of the regeneration compared to those in the wild-type protoplasts (Fig. 3D).
Figure 3.
Expression levels of purple acid phosphatase (PAP) and acid phosphatase activities in the transgenic cells overexpressing NtPAP12 and in the wild-type cells. SDS-PAGE under nonreducing conditions was performed on 20 μg of wall proteins prepared from cells at a logarithmic growth phase (A) or from protoplasts regenerating walls for 30 min (B). In each case, immunoblot analysis was performed using rabbit anti-wall-bound purple acid phosphatase antibody. C and D, Acid phosphatase activity in the walls from suspension-cultured tobacco cells at the exponential phase (C) or from protoplasts at the early stage of wall regeneration (D). White circles, Wild-type protoplasts; black circles, transgenic protoplasts overexpressing NtPAP12. The activity values represent the mean of three incubation mixtures for each reaction with individual values varying from the mean by <5%.
β-Glucan formed at the surface of the protoplasts overexpressing NtPAP12 as judged by Calcofluor White staining (Supplemental Fig. S1). The amount of polysaccharide synthesized by the transgenic protoplasts after 60 min regeneration were about 2-fold higher than in the case of the wild-type protoplasts (Table I). Based on methylation analysis, terminal, 3-linked, and 4-linked Glcs were each increased in the regenerated walls of the transgenic cells compared to wild-type cells, by factors of 1.8, 2.4, and 1.7, respectively (Table II). This indicates that the expression of the phosphatase was accompanied by enhanced β-glucan deposition, during which β-glucan synthases might be activated. In suspension-cultured cells the overexpression of NtPAP12 provoked an increase of cellulose deposition during cell growth compared to the wild-type cells, while the pectin content was slightly decreased (Supplemental Fig. S2). Altogether these results demonstrate that overexpression of NtPAP12 increased callose and cellulose deposition in tobacco cells.
Table I.
Amount of extracellular polysaccharides formed by protoplasts at 60 h after onset of wall regeneration
Total polysaccharides of cell walls isolated from wild-type and transgenic protoplasts, and total polysaccharides secreted by wild-type and transgenic protoplasts into the cell wall regeneration medium were determined by the phenol-sulfuric acid method (see “Materials and Methods”). Average and sd values were obtained from three different experiments. PAP, Purple acid phosphatase.
| Fraction | Wild Type | PAP Overexpression |
|---|---|---|
| μg per mL cells | ||
| Wall | 32 ± 4 | 61 ± 5 |
| Medium | 23 ± 2 | 52 ± 4 |
Table II.
Sugar linkages of polysaccharides from the regenerated walls of protoplasts
Glycosyl linkages were determined as methylated alditolacetates by gas-liquid chromatography. Average and sd values were obtained from three different experiments. PAP, Purple acid phosphatase.
| Glycosyl Linkage | Wild Type | PAP Overexpression |
|---|---|---|
| nmol per mL cells | ||
| Terminal Ara | 10.5 ± 0.3 | 4.4 ± 0.1 |
| Terminal Xyl | 2.6 ± 0.1 | 2.1 ± 0.1 |
| Terminal Glc | 17.6 ± 0.6 | 31.7 ± 0.9 |
| 5-linked Ara | 9.2 ± 0.2 | 9.5 ± 0.5 |
| 4-linked Xyl | 18.4 ± 0.6 | 12.7 ± 0.6 |
| 3-linked Glc | 8.8 ± 0.4 | 21.1 ± 0.8 |
| 4-linked Man | 11.0 ± 0.5 | 7.3 ± 0.3 |
| 3-linked Gal | 9.9 ± 0.4 | 9.1 ± 0.4 |
| 4-linked Glc | 86.8 ± 2.4 | 145.6 ± 5.2 |
| 6-linked Gal | 5.7 ± 0.2 | 5.5 ± 0.3 |
| 4,6-linked Glc | 6.5 ± 0.2 | 5.3 ± 0.2 |
| 3,6-linked Gal | 13.1 ± 0.6 | 14.1 ± 0.6 |
Effect of Phosphatase Overexpression on in Vitro β-Glucan Synthase Activities
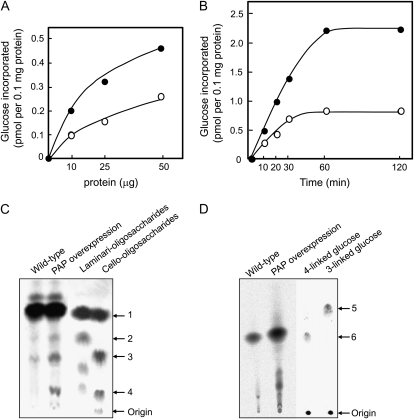
To further investigate whether the overexpression of NtPAP12 is accompanied by higher callose and cellulose synthase activities in vitro, we measured the activity of glucan synthases in digitonin extracts of microsomal preparations from transgenic and wild-type tobacco cells. Glucan synthase activities increased with increasing amounts of membrane proteins in the assay mixture for both enzyme preparations (Fig. 4A). However, after 10 min incubation of the detergent extracts in the presence of UDP-Glc, the incorporation of Glc into glucans was twice as high in the case of the transgenic cells at any protein concentration tested (Fig. 4A). The Glc incorporation was also systematically higher during the time course in the case of the detergent extracts from the transgenic cells (Fig. 4B). In particular, glucan formation from enzyme extracts from the transgenic cells was about 2-fold higher during the initial phase of incorporation and 2.7-fold higher at the extent of incorporation, compared to wild-type cells.
Figure 4.
In vitro synthesis of β-glucans and product characterization. A, Reaction performed in the presence of variable amounts of protein during 10 min in conditions favoring cellulose biosynthesis (see “Materials and Methods”). White circles, Wild type; black circles, cells overexpressing purple acid phosphatase (PAP). The values represent the mean of three incubation mixtures for each reaction with individual values varying from the mean by <5%. B, Time course synthesis of β-glucan. Ten micrograms of protein was used for each reaction. White circles, wild type; black circle, cells overexpressing purple acid phosphatase. The values represent the mean of three incubation mixtures for each reaction with individual values varying from the mean by <5%. C, Paper chromatography of the enzymatic hydrolysates (cellulase) corresponding to the β-glucans synthesized in vitro in conditions favoring cellulose biosynthesis. 14C-Polysaccharides formed from UDP-[14C]Glc were precipitated with 70% ethanol containing 1 mm EDTA and washed four times with ethanol. Radioactive products (278 Bq) were used for the wild-type and the transgenic cells. Arrows 1, 2, 3, and 4 indicate the position of Glc, laminaribiose, cellobiose, and cellotriose, respectively. D, Thin-layer chromatography of the methylated alditol acetates corresponding to the glucans synthesized in vitro. Radioactive products treated by the acetic/nitric acid reagent were used for the wild-type (56 Bq) and the transgenic cells (111 Bq). Arrows 5 and 6 indicate 3-linked Glc and 4-linked Glc, respectively.
To confirm the nature of the reaction products, the insoluble material obtained from the in vitro synthesis experiment was subjected to enzymatic hydrolysis with a cellulase and the hydrolysate was analyzed by paper chromatography. The analysis revealed that the hydrolysate corresponding to the glucans from the transgenic cells contained significantly higher amounts of cellobiose and cellotriose compared to the situation in wild-type cells (Fig. 4C). Laminaribiose was also faintly observed in the hydrolysates. In addition, the overall amount of glucan synthesized was significantly higher for the transgenic cells than for the wild-type cells, thereby confirming the results obtained prior to product characterization (Fig. 4, A and B). Altogether these results showed that the phosphatase significantly accelerated the synthesis of β-1,4-glucan together with callose.
The glucan products after treatment with the acetic acid/nitric acid (Updegraff) reagent (Updegraff, 1969) were also subjected to methylation analysis and the resulting permethylated alditol acetates were analyzed by paper chromatography (Fig. 4D). The alditol acetate characteristic of 4-linked Glc (1,4,5-tri-O-acetyl-2,3,6-tri-O-methyl-d-glucitol) was detected in the preparations obtained from both the wild-type and the transgenic cells. The intensity of the spot was about twice as high in the case of the product from the transgenic cells compared to that of the wild-type cells (Fig. 4D). This confirms a higher synthesis of cellulose in the transgenic cells. Altogether these data indicated that cellulose synthase activity was higher in the cells overexpressing NtPAP12.
To further analyze the extent of activation of glucan synthases from different types of membranes, we fractionated the microsomal fraction into intracellular and plasma membranes and measured glucan synthase activities in preparations from both the wild-type and the transgenic cells (Table III). In agreement with the data presented above, the activity of callose synthase, which is located in the plasma membrane and catalyzes the incorporation of Glc from UDP-Glc into 1,3-β-glucan (glucan synthase II), was 1.8-fold higher in the preparation from the transgenic cells than in the preparation from wild-type cells. However, no difference in activity was observed for the Golgi-located 1,4-β-glucan synthase (glucan synthase I) or xyloglucan xylosyltransferase, which shows that the overexpression of NtPAP12 did not affect glycan synthases from the Golgi membranes. The phosphatase instead promotes the activities of callose and cellulose synthases (Tables II and III; Fig. 4), which are located in the plasma membrane.
Table III.
β-Glucan synthase activities in the Golgi and plasma membranes fractionated from total microsomal fractions
Average and sd values were obtained from three different experiments. PAP, Purple acid phosphatase.
| Plant | Golgi-Bound 1,4-β-Glucan Synthase | Plasma Membrane-Bound Callose Synthase | Xyloglucan Xylosyltransferase |
|---|---|---|---|
| pmol mg−1 protein | |||
| Wild type | 38.3 ± 0.9 | 4,183 ± 133 | 30.8 ± 0.8 |
| PAP overexpression | 37.7 ± 1.2 | 7,652 ± 201 | 28.8 ± 1.1 |
DISCUSSION
Our data demonstrate that the activities of callose and cellulose synthases are enhanced by the overexpression of the wall-bound purple acid phosphatase in tobacco cells. This is supported by several lines of evidence: (1) the transgenic protoplasts regenerating a cell wall exhibit a higher rate of β-glucan synthesis; (2) the higher level of phosphatase activity in the walls of the transgenic cells is correlated with a higher level of glucan synthase activities in detergent extracts of microsomal fractions; (3) the higher glucan synthase activities observed in the transgenic line are specific for the plasma membrane, whereas no activation of Golgi-bound glycan synthases was detected; and (4) product characterization based on enzymatic hydrolysis and methylation analysis unequivocally demonstrates that the glucans synthesized in vitro by the detergent extracts from both the wild-type and transgenic cells are callose and cellulose, and that the amount of each glucan synthesized by the enzyme preparation from the transgenic cells is higher than that synthesized by the fraction from the wild-type cells. It seems likely that callose synthase activity is regulated by phosphorylation/dephosphorylation. It remains, however, to determine whether the phosphorylation event occurs directly on the callose synthase catalytic subunit or on another protein that itself controls callose synthase activity. A number of proteins have been identified to be potentially involved in the regulation of callose synthases; they could be the target of NtPAP12. These include phragmoplastin, the GTP-binding protein Rop1, and a UDP-glucosyltransferase potentially associated to the callose synthase complex (Hong et al., 2001a, 2001b). However, the localization of Rop1 on the cytoplasmic side of the plasma membrane is not compatible with the localization of the product of the NtPAP12 gene, which is located in the cell wall. In addition, phragmoplastin is thought to interact only with the callose synthase complex responsible for cell plate formation (Hong et al., 2001a) while our data suggest a general activation of callose synthase activity in protoplasts regenerating a cell wall. Similarly, in the case of cellulose synthase, the phosphorylation/dephosphorylation process may occur directly on the catalytic subunit itself. This is supported by the identification of several phosphorylation sites in cellulose synthases from Arabidopsis (Nühse et al., 2004). However, these sites were predicted to be located on the cytoplasmic side of the plasma membrane, which is not compatible with the cell wall localization of NtPAP12. The identification of phosphorylation sites in cellulose synthase was also reported for AtCesA7, which is involved in secondary cell wall formation in Arabidopsis (Taylor, 2007). The data point toward a role of phosphorylation in regulating the turnover of cellulose synthase by proteolysis through a proteasome-dependent pathway (Taylor, 2007), which again implies a cytoplasmic phosphorylation event. Thus, the information available to date combined with our present data suggest that the wall-bound NtPAP12 phosphatase from tobacco is involved in the regulation of the cellulose synthase activity by acting on an unidentified membrane protein, which may lead to the promotion of cellulose synthesis. NtPAP12 was shown to be secreted to the wall via the classical secretion pathway involving Golgi vesicles (Sano et al., 2003; Kaida et al., 2008). Since the levels of Golgi-located glycosyltransferases were not affected in the tobacco cells overexpressing NtPAP12, it can be hypothesized either that NtPAP12 is not functional in the Golgi but only in its final destination, i.e. the cell wall, or that the Golgi-bound glycosyltransferases are not regulated by phosphorylation. Further work is needed to demonstrate whether NtPAP12 is involved in the coregulation of callose and cellulose synthases as suggested earlier (Delmer, 1999).
Arabidopsis contains a large family of purple acid phosphatases, which is composed of 29 genes, 28 of which have signal peptides that are potentially involved in the transfer of the proteins to the wall and/or vacuole. As the amino acid sequence of NtPAP12 is most similar to that of AtPAP10 (amino acid identity higher than 80%), the analysis of the corresponding knockout plants is in progress in our laboratory to further characterize the function of these proteins.
MATERIALS AND METHODS
Cell Cultures
The tobacco (Nicotiana tabacum) cell line XD-6 derived from tobacco var. Xanthi and the transgenic cells overexpressing NtPAP12 (Kaida et al., 2003) under the S35 constitutive promoter were cultured in 40 mL of Murashige and Skoog medium (Murashige and Skoog, 1962) containing 3% Suc and 4.5 μm 2,4-dichlorophenoxyacetic acid in 100-mL flasks. The flasks were shaken at 110 rpm at 25°C for 10 d.
Transformation of Tobacco Cells
NtPAP12 was amplified from tobacco cDNA (Kaida et al., 2003) by PCR using a forward primer (5′-CTGGATCCTGGTGTGCAAAAATGGGTGTG-3′) and reverse primer (5′-TTCACGATTTGGTTACCATGGACTC-3′). Next, the cDNA was digested with BamHI and SacI, then introduced into the binary vector under the control of an enhanced cauliflower mosaic virus 35S promoter (Mitsuhara et al., 1996), and the plasmid was transformed into Agrobacterium tumefaciens LBA4404.
The transformation of tobacco cells was performed according to the method described previously (An, 1985). Tobacco cells (cell line XD-6) were cocultivated with the bacteria for 4 d at 26°C. Then the cells were cultured in 40 mL of Murashige and Skoog medium supplemented with 50 μg/mL kanamycin and 500 μg/mL cefotaxime. When the culture reached the stationary phase, 3 mL of culture was transferred to fresh medium supplemented with 200 μg/mL kanamycin and 500 μg/mL cefotaxime. After an additional 10 culture cycles in the selective medium, we established a transgenic cell line that had been cultured more than 200 times in the presence of 50 μg/mL kanamycin. During inoculation, the phosphatase activity was always determined using phosphotyrosine as a substrate. The specific activity of phosphatase was always more than 20-fold higher for the transgenic cells (0.3 unit/mg total proteins) than for the wild-type cells (0.014 unit/mg total proteins) over a period of 5 years. All of the following experiments were performed using a culture medium devoid of antibiotics.
Preparation and Culture of Protoplasts for Wall Regeneration
Protoplasts were obtained by digestion of cell walls of suspension-cultured tobacco cells with 1.5% (w/v) cellulase (Cellulase Onozuka RS, Yakult) and 0.05% (w/v) pectolyase Y-23 (Seishin Pharmaceutical) in the presence of 0.48 m mannitol at pH 5.2 for 40 min. The protoplasts were filtered through a 60-μm nylon mesh and purified by sedimentation through 13.7% Suc solution at 170g for 1 min. Then, they were washed with 0.48 m mannitol and cultured in a medium for cell wall regeneration (Nagata and Takebe, 1970) at a density of 105 protoplasts mL−1 at 26°C, and harvested by centrifugation at 170g for 1 min. The cell wall regeneration medium (pH 5.8) contained 20 mm CaCl2, 0.2 mm KH2PO4, 1.0 mm KNO3, and 2.0 mm MgSO4 as major component, 1.0 μm KI and 0.01 μm CuSO4 as minor component, 1.0 μg L−1 thiamine, 100 μg L−1 inositol, 2.0 μg L−1 Gly, and 0.48 m mannitol.
Preparation of Plasma Membrane Sheets and Immunofluorescence Staining
Localizations of extracellular purple acid phosphatase and β-glucans were determined using plasma membrane sheets (Hirai et al., 1998). The protoplasts were immediately transferred to a coverslip that had been coated with poly-l-Lys (molecular weight 70,000–150,000, Sigma-Aldrich), and covered with the medium for cell wall regeneration. After incubation, the protoplasts were subjected to lysis in 50 mm PIPES-KOH (pH 6.8) containing 1 mm MgCl2 and 5 mm EGTA, then fixed with 50 mm PIPES-KOH (pH 6.8) containing 3.7% (w/v) formaldehyde, 1 mm MgSO4, 5 mm EDTA, and 1% glycerol. Membrane permeabilization was induced by treatment with 50 mm PIPES-KOH (pH 6.8) containing 0.5% (v/v) Triton X-100, 1 mm MgSO4, 5 mm EDTA, and 1% glycerol for 5 min. Prior to immunostaining, the membrane sheets were blocked with phosphate-buffered saline (20 mm sodium phosphate, pH 7.0 and 150 mm NaCl) containing 1% (w/v) bovine serum albumin, 0.1 m Gly, and 0.05% Triton X-100 for 20 min. Purple acid phosphatase was detected with a rabbit antitobacco purple acid phosphatase antibody and Alexa Fluor 568 conjugated antirabbit IgG (Invitrogen). β-Glucan microfibrils were stained with 0.002% Calcofluor White. The membrane sheets were subsequently embedded in an antifading reagent (SlowFade Light; Invitrogen) and observed under a fluorescence microscope (Olympus BX50) equipped with a confocal laser-scanning head system (Leica TCSNT).
Preparation of Purple Acid Phosphatase
Suspension-cultured tobacco cells were homogenized in 50 mm HEPES-KOH (pH 7.0) buffer containing 1 mm dithiothreitol, using a Teflon-glass homogenizer at 1,500 rpm for 20 min. The wall materials were collected on nylon mesh (pore size, 50 μm) and washed with 10 mm HEPES-KOH buffer (pH 7.0) by centrifugation at 5,000g for 20 min. The wall-bound phosphatase was extracted with the same buffer containing 0.7 m NaCl. The extract was concentrated by precipitation with ammonium sulfate at 65% saturation. Following centrifugation at 14,000g for 30 min, the pellet was dissolved with 10 mm HEPES-KOH buffer (pH 7.0) and dialyzed against the same buffer.
The protoplasts cultured in the medium for cell wall regeneration were harvested by centrifugation at 500g for 2 min and homogenized in 50 mm HEPES-KOH (pH 7.0) containing 1 mm dithiothreitol using a Teflon-glass homogenizer at 1,200 rpm for 3 min. The homogenate was overlaid on 50 mm HEPES-KOH (pH 7.0) containing 40% Suc and centrifuged at 40,000g for 30 min. The wall fraction (pellet) was collected and washed with 10 mm HEPES-KOH buffer (pH 7.0) by centrifugation at 10,000g for 15 min. The wall-bound phosphatase was extracted with the same buffer containing 0.7 m NaCl.
Immunoblotting Analysis
Twenty micrograms each of wall-bound proteins prepared as described above were separated by SDS-PAGE under nonreducing conditions. The proteins were electrotransferred to Hybond-ECL (GE Healthcare) and probed first with rabbit anti-wall-bound purple acid phosphatase antibody, then with a peroxidase-conjugated secondary antibody. Detection was performed using ECL western blotting detection reagents (GE Healthcare).
Assay for Phosphatase
Phosphatase activity was assayed in a reaction mixture containing the enzyme preparation, 1 mm phosphotyrosine, and 50 mm sodium acetate buffer (pH 5.8) in a total volume of 0.1 mL. The phosphate released over the course of 3 min at 30°C was assayed according to Van Veldhoven and Mannaerts (1987). One unit of activity was defined as the amount of enzyme necessary to produce 1 μmol of phosphate in 1 min at 30°C.
Fractionation of Wall Polysaccharides from Tobacco Cells and Protoplasts
Tobacco cells were homogenized in 50 mm HEPES-KOH (pH 7.0) containing 1 mm dithiothreitol using a Teflon-glass homogenizer at 1,400 rpm for 20 min. The homogenates were filtered on a nylon mesh (60 μm). The residue on the nylon mesh was washed five times with 10 mm HEPES-KOH (pH 7.0; centrifugation at 1,000g for 15 min).
The wall fractions were boiled for 3 min and treated with salivary amylase and proteinase K (GibcoBRL). The regenerated wall fractions were subjected to methylation analysis as described below. Carbohydrate contents recovered from growing cells were determined after cell wall fractionation as follows: Pectin and hemicellulose were extracted sequentially five times with 0.1 m EDTA (pH 7.0) at 85°C and five times with 24% (w/w) KOH containing 0.1% NaBH4 at 45°C, respectively. The final insoluble residue corresponding to cellulose was dissolved with ice-cold 72% (w/v) sulfuric acid. Total sugar in each fraction was determined by the phenol-sulfuric acid method (Dubois et al., 1956).
Preparation of Microsomal Membranes
The wild-type and transgenic cells (50 g) were homogenized in a mortar using 50 mL of homogenization buffer (50 mm MES-KOH, pH 6.0, containing 1 mm dithiothreitol, 1 mm EDTA, 0.4 m Suc, 10 mm KCl, and 2% bovine serum albumin). The homogenates were centrifuged at 5,000g for 10 min. The supernatant was layered onto 20 mm MES-KOH, pH 6.0, containing 1 mm dithiothreitol and 1.6 m Suc (1.6 m Suc buffer) and centrifuged at 200,000g for 20 min. Microsomal membranes were observed as a thin band between the homogenization buffer and the 1.6 m Suc buffer. The membrane fractions were suspended in 20 mm MES-KOH, pH 6.0 containing 1 mm dithiothreitol, 1 mm EDTA, and 0.4 m Suc, again layered on a 1.6 m Suc buffer, and centrifuged at 200,000g for 20 min. About 1 mL of microsomal membranes was collected. The microsomal membranes from the wild-type cells (4.5 mg protein/mL) and those from the transgenic cells (5.7 mg protein/mL) were subjected to glucan synthase assays.
Preparation of Detergent Extracts from Microsomal Membranes and Assays for Glucan Synthases
Digitonin extracts were prepared from the microsomal fraction as described elsewhere (Colombani et al., 2004). Cellulose synthase was measured by the method of Lai-Kee-Him et al. (2002). The detergent extracts from microsomal membranes (50 μg of protein/150 μL) were incubated with a mixture of UDP-Glc and UDP-[14C]Glc (6.1 GBq/mol; 1 mm total final concentration), 10 mm MgCl2, 2 mm cellobiose, and 20 mm HEPES-KOH (pH 6.8) in a total volume of 200 μL. After incubation for 10 min at 25°C, the reaction was stopped by the addition of the Updegraff reagent (80% acetic acid/concentrated nitric acid, 10:1; Updegraff, 1969). The mixture was placed in a boiling water bath for 30 min and the resulting insoluble material was washed three times with water and 1 m acetic acid on disc-glass filters by filtration, and the radioactivity remaining on the filters was measured by liquid scintillation. The proportion of callose and cellulose synthesized in these conditions were determined as described below (see “Sugar Analysis”).
Callose synthase in the fractions recovered after fractionation of the microsomal fraction into Golgi and plasma membranes was measured by the method of Van der Woude et al. (1974). The incubation mixture contained 2 μm UDP-[14C]Glc (12.1 GBq/mmol), 1 mm MgCl2, 0.1 mm cellobiose, 20 mm HEPES-KOH (pH 6.8), and the enzyme preparation (100 μg of protein) in a total volume of 20 μL. After incubation for 15 min at 25°C, the reaction was stopped by the addition of 1 m NaOH. The insoluble material was washed three times with water and 1 m acetic acid on glass filters by filtration and the corresponding radioactivity was measured by liquid scintillation.
Golgi-bound 1,4-β-glucan synthase was measured by the method of Ordin and Hall (1968). The mixture contained 2 mm UDP-[14C]Glc (12.1 GBq/mol), 0.1 mm cellobiose, 20 mm HEPES-KOH (pH 6.8), and the enzyme preparation (100 μg of protein) in a total volume of 20 μL. After incubation for 15 min at 25°C, the reaction was stopped by the addition of 1 m NaOH. The insoluble material was washed three times with water and 1 m acetic acid on disc-glass filters by filtration, and the radioactivity retained on the filters was measured.
Xyloglucan 6-α-d-xylosyltransferase activity was determined as described previously (Hayashi et al., 1988). The incubation mixture contained 2 μm UDP-[14C]Xyl (9.7 GBq/mmol), 1 mm MnCl2, 20 mm HEPES-KOH (pH 6.8), and the enzyme preparation (100 μg of protein) in a total volume of 20 μL. After incubation for 15 min at 25°C, the reaction was stopped by heating in a boiling water bath for 5 min. Fifty microliters of carrier soybean (Glycine max) xyloglucan solution containing 100 μg of xyloglucan and 1 mL of 95% ethanol were added to the mixture, and the mixture was stirred and centrifuged. The supernatant was removed, and the precipitate was washed with 70% ethanol. The precipitate was suspended in 1 mL of 50 mm potassium acetate buffer (pH 5.0) containing 1 mg of Aspergillus oryzae enzyme preparation, and incubated for 12 h at 40°C. The reaction was stopped by heating. The mixture was then deionized with Amberlite IR-120 (H+) and concentrated to dryness in vacuo. The digest was analyzed by paper chromatography with 1-propanol/ethyl acetate/water, 3:2:1, as solvent. The area corresponding to isoprimeverose on the paper was cut out and its radioactivity was measured. Units of specific activity were defined as the amounts of [14C]Xyl (pmol) incorporated into isoprimeverose per 15 min, per mg protein.
Sugar Analysis
14C-Polysaccharides formed from UDP-[14C]Glc were digested with Streptomyces griceus endocellulase in 20 mm of sodium acetate buffer (pH 5.5) at 40°C for 48 h. The mixture was then deionized with Amberlite IR-120 (H+) and concentrated to dryness in vacuo. The digest was analyzed by paper chromatography with 1-propanol/ethyl acetate/water, 6:1:2, as solvent.
Methylation analysis was carried out as described previously (Hayashi, 1989). Polysaccharides (100 μg) of wall fractions were dissolved in 0.5 mL dimethyl sulfoxide and methyl sulfinyl carbanion (0.5 mL) was added to the mixture. The mixture was agitated in an ultrasonic bath at 45°C for 4 h. Methyl iodide (0.5 mL) was added dropwise during external cooling in ice water. The mixture was again agitated in the ultrasonic bath for 30 min, dialyzed against tap water overnight to remove dimethyl sulfoxide, and concentrated to dryness. The methylated polysaccharide was dissolved in 1.5 mL of 2 m trifluoroacetic acid and hydrolyzed for 1 h at 120°C. The mixture was concentrated to dryness and 1 mL of 0.5 m NaBH4 in 2 m NH4OH was added. After reduction at 60°C for 1 h, the resulting alditols were acetylated according to the method of Blakeney et al. (1983). The partially methylated alditol acetates were separated with a gas chromatography system (Agilent Technologies) using a glass capillary column DB-225 (0.25 mm × 15 m, Agilent Technologies) with temperature rising linearly from 140°C to 190°C at a rate of 2°C min−1. The peaks were identified by mass spectrometry (JMS K9, JEOL). The radioactive acetates were also separated by thin-layer chromatography on silica gel at 50°C for 80 min using 1-hexane/diethyl ether/acetic acid, 6:3:1, as solvent.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AB017967 (NtPAP12).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Wall regeneration in tobacco protoplasts overexpressing purple acid phosphatase.
Supplemental Figure S2. Carbohydrate contents of the cell walls recovered from growing cell suspension cultures.
Supplementary Material
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI (grant nos. 19208016 and 19405030), by the Swedish Centre for Biomimetic Fibre Engineering (Biomime), and by a grant from the JSPS (to V.B.). This work is also part of the outcome of the JSPS Global COE Program (E-04): In Search of Sustainable Humanosphere in Asia and Africa.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Takahisa Hayashi (taka@rish.kyoto-u.ac.jp).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- An G (1985) High efficiency transformation of cultured tobacco cells. Plant Physiol 79 568–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279 717–720 [DOI] [PubMed] [Google Scholar]
- Blakeney AB, Harris PJ, Henry RJ, Stone BA (1983) Simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr Res 113 291–299 [Google Scholar]
- Brownfield L, Ford K, Doblin MS, Newbigin E, Read S, Bacic A (2007) Proteomic and biochemical evidence links the callose synthase in Nicotiana alata pollen tubes to the product of the NaGSL1 gene. Plant J 52 147–156 [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37 D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani A, Djerbi S, Bessueille L, Blomqvist K, Ohlsson A, Berglund T, Teeri TT, Bulone V (2004) In vitro synthesis of (1→3)-β-d-glucan (callose) and cellulose by detergent extracts of membranes from cell suspension cultures of hybrid aspen. Cellulose 11 313–327 [Google Scholar]
- Cui X, Shin H, Song C, Laosinchai W, Amano Y, Brown RM Jr (2001) A putative plant homolog of the yeast (1→3)-β-glucan synthase subunit FKS1 from cotton (Gossypium hirsutum L.) fibers. Planta 213 223–230 [DOI] [PubMed] [Google Scholar]
- Delmer DP (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50 245–276 [DOI] [PubMed] [Google Scholar]
- Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM (2001) Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol 125 2040–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton LK, Rebers PA, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28 350–356 [Google Scholar]
- Hayashi T (1989) Measuring β-glucan deposition in plant cell walls. In HF Linskens, JF Jackson, eds, Modern Methods of Plant Analysis, Plant Fibers 10. Springer-Verlag, Berlin, pp 138–160
- Hayashi T, Koyama T, Matsuda K (1988) Formation of UDP-xylose and xyloglucan in soybean Golgi membranes. Plant Physiol 87 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai N, Sonobe S, Hayashi T (1998) In situ synthesis of β-glucan microfibrils on tobacco plasma membrane sheets. Proc Natl Acad Sci USA 95 15102–15106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Delauney AJ, Verma DPS (2001. a) A cell-plate specific callose synthase and its interaction with phragmoplastin. Plant Cell 13 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Zhang Z, Olson JM, Verma DPS (2001. b) A novel UDP-Glc transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 13 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida R, Hayashi T, Kaneko T (2008) Purple acid phosphatase in the walls of tobacco cells. Phytochemistry 69 2546–2551 [DOI] [PubMed] [Google Scholar]
- Kaida R, Sage-Ono K, Kamada H, Okuyama H, Syono K, Kaneko TS (2003) Isolation and characterization of four cell wall purple acid phosphatase genes from tobacco cells. Biochim Biophys Acta 1625 134–140 [DOI] [PubMed] [Google Scholar]
- Lai-Kee-Him J, Chanzy H, Müller M, Putaux JL, Imai T, Bulone V (2002) In vitro versus in vivo cellulose microfibrils from plant primary wall synthases: structural differences. J Biol Chem 277 36931–36939 [DOI] [PubMed] [Google Scholar]
- Li J, Burton RA, Harvey AJ, Hrmova M, Wardak AZ, Stone BA, Fincher GB (2003) Biochemical evidence linking a putative callose synthase gene with (1→3)-β-d-glucan biosynthesis in barley. Plant Mol Biol 53 213–225 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37 49–59 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15 473–497 [Google Scholar]
- Nagata T, Takebe I (1970) Cell wall regeneration and cell division in isolated tobacco mesophyll protoplast. Planta 92 301–308 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Stensballe A, Jensen ON, Peck SC (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordin L, Hall MA (1968) Cellulose synthesis in higher plants from UDP Glc. Plant Physiol 43 473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliyath G, Poovaiah BW (1988) Promotion of β-glucan synthesis activity in corn microsomal membranes by calcium and protein phosphorylation. Plant Cell Physiol 29 67–73 [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PM (1973) Regulation of β-glucan synthetase activity by auxin in pea stem tissue. II. Metabolic requirements. Plant Physiol 51 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano A, Kaida R, Maki H, Kaneko TS (2003) Involvement of an acid phosphatase on cell wall regeneration of tobacco protoplast. Physiol Plant 119 121–125 [Google Scholar]
- Shea EM, Gibeaut DM, Carpita NC (1989) Structural analysis of the cell walls regenerated by carrot protoplasts. Planta 179 293–308 [DOI] [PubMed] [Google Scholar]
- Taylor NG (2007) Identification of cellulose synthase AtCesA7 (IRX3) in vivo phosphorylation sites—a potential role in regulating protein degradation. Plant Mol Biol 64 161–171 [DOI] [PubMed] [Google Scholar]
- Updegraff DM (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32 420–424 [DOI] [PubMed] [Google Scholar]
- Van der Woude WJ, Lembi CA, Morre DJ, Kindinger JI, Ordin L (1974) β-Glucan synthetases of plasma membrane and Golgi apparatus from onion stem. Plant Physiol 54 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veldhoven PP, Mannaerts GP (1987) Inorganic and organic phosphate measurements in the nanomolar range. Anal Biochem 161 45–48 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.