Abstract
The pepper (Capsicum annuum) bacterial spot (Bs) resistance gene Bs3 and its allelic variant Bs3-E mediate recognition of the Xanthomonas campestris pv vesicatoria type III effector protein AvrBs3 and its deletion derivative AvrBs3Δrep16. Recognition specificity resides in the Bs3 and Bs3-E promoters and is determined by a defined promoter region, the UPA (for up-regulated by AvrBs3) box. Using site-directed mutagenesis, we defined the exact boundaries of the UPAAvrBs3 box of the Bs3 promoter and the UPAAvrBs3Δrep16 box of the Bs3-E promoter and show that both boxes overlap by at least 11 nucleotides. Despite partial sequence identity, the UPAAvrBs3 box and the UPAAvrBs3Δrep16 box were bound specifically by the corresponding AvrBs3 and AvrBs3Δrep16 proteins, respectively, suggesting that selective promoter binding of AvrBs3-like proteins is the basis for promoter activation specificity. We also demonstrate that the UPAAvrBs3 box retains its functionality at different positions within the pepper Bs3 promoter and confers AvrBs3 inducibility in a novel promoter context. Notably, the transfer of the UPAAvrBs3 box to different promoter locations is always correlated with a new transcriptional start site. The analysis of naturally occurring Bs3 alleles revealed many pepper accessions that encode a nonfunctional Bs3 variant. These accessions showed no apparent abnormalities, supporting the supposition that Bs3 functions only in disease resistance and not in other developmental or physiological processes.
Plant pathogenic microbes deliver a cocktail of effector proteins into the host cytoplasm that collectively promote microbial growth (Kamoun, 2006; Block et al., 2008; Göhre and Robatzek, 2008; Cunnac et al., 2009; Hogenhout et al., 2009). Many effector proteins were first termed avirulence (Avr) proteins because their presence evoked a hypersensitive response (HR) in plants expressing a matching resistance (R) gene (Grant et al., 2006; Bent and Mackey, 2007). Although the appearance of an HR often correlates with disease resistance, its causal role in plant immunity has not been fully elucidated (Greenberg and Yao, 2004). Avr proteins were identified initially as activators of the plant immune reaction, but many were later found to contribute to pathogen virulence on host plants that lack a corresponding R gene (Jones and Dangl, 2006). Meanwhile, the in planta function of a number of effectors has been studied at the molecular level, and some seem to function as enzymes (Mudgett, 2005; Abramovitch et al., 2006; Chisholm et al., 2006). Yet, some effectors of the bacterial genus Xanthomonas harbor nuclear localization signals and a transcriptional activation domain (Van den Ackerveken et al., 1996; Yang et al., 2000; Szurek et al., 2001) and have been termed transcription activator-like effector proteins (TALes; Kay and Bonas, 2009). The prototype TALe, AvrBs3, was identified from Xanthomonas campestris pv vesicatoria (Xcv) based on its avirulence activity in pepper (Capsicum annuum; Bonas et al., 1989) and was later shown to contribute to bacterial virulence in susceptible pepper genotypes (Wichmann and Bergelson, 2004). The most characteristic structural feature of TALes is a variable number of tandemly arranged, nearly perfect copies of a 34-amino acid motif that mediates binding of the TALe AvrBs3 to host target promoters (Kay et al., 2007).
Although TALes are generally highly homologous to each other, their activity in plants is subject to exquisite specificity (Schornack et al., 2006). For example, the pepper Bs3 and the tomato (Solanum lycopersicum) Bs4 R genes mediate recognition of the 96.6% identical Xcv AvrBs3 and AvrBs4 proteins, respectively (Ballvora et al., 2001; Schornack et al., 2005; Römer et al., 2007). Tomato Bs4 is expressed constitutively at low levels and encodes a nucleotide-binding Leu-rich repeat-type R protein (Schornack et al., 2004, 2005). By contrast, the pepper Bs3 gene is transcriptionally induced by AvrBs3 and encodes a YUCCA-like flavin monooxygenase (Römer et al., 2007). Thus, Bs3- and Bs4-mediated recognition are mechanistically distinct despite the fact that they mediate recognition of almost identical effector proteins.
Previously, we showed that the pepper Bs3 gene mediates recognition of AvrBs3 but not of its deletion derivative AvrBs3Δrep16, which lacks repeat units 11 to 14 (Herbers et al., 1992). Reciprocally, the Bs3-E allele mediates recognition of AvrBs3Δrep16 but not of AvrBs3. Recent studies demonstrated that AvrBs3 and AvrBs3Δrep16 specifically activate the matching Bs3 and Bs3-E promoters, respectively (Römer et al., 2007). The Bs3-E gene is an allele of Bs3 that carries a 13-bp insertion in its promoter compared with Bs3. Comparison of the Bs3 and other AvrBs3-inducible promoters from pepper revealed a conserved DNA element, the so-called UPA (for up-regulated by AvrBs3) box (Kay et al., 2007; Römer et al., 2007). Notably, the Bs3-E-specific 13-bp insertion is located within the UPA box of the Bs3 promoter. Electrophoretic mobility shift assays (EMSA) showed that AvrBs3 has a higher affinity to the Bs3 promoter as compared with the Bs3-E promoter. Yet, in EMSA, AvrBs3Δrep16 also had a higher affinity for the Bs3 promoter than for the Bs3-E promoter. Thus, it seemed that promoter binding of AvrBs3 or AvrBs3Δrep16 is not the basis for promoter activation specificity (Römer et al., 2007).
In order to gain further insight into the molecular basis of TALe specificity and Bs3-mediated resistance, we have now carried out site-directed mutagenesis to define the exact boundaries of the UPA boxes of the Bs3 promoter (herein designated UPAAvrBs3 box) and the Bs3-E promoter (herein designated UPAAvrBs3Δrep16 box). We present new EMSA results that demonstrate that promoter binding is indeed the basis for promoter activation specificity by TALes. Finally, the analysis of a collection of naturally occurring Bs3 alleles revealed that pepper accessions encoding nonfunctional Bs3 variants are phenotypically normal. These data support the role of Bs3 exclusively in disease resistance.
RESULTS
Mapping the Promoter Sequences Used for Activation by AvrBs3 and AvrBs3Δrep16
Previously, we showed that transcript abundance of the pepper Bs3 resistance gene is increased 24 h post infection with Xcv strains that deliver AvrBs3 (Römer et al., 2007). We have now carried out semiquantitative reverse transcription (RT)-PCR to monitor Bs3 transcript accumulation on Xcv-infected leaves in a time-course experiment. As shown in Figure 1A, Bs3 transcript was detectable as early as 6 h post infection and peaks at 18 h post infection. Expression of Bs3 and Bs3-E was also studied in uninfected leaf, flower, fruit, and root tissue. However, we were unable to detect Bs3 or Bs3-E transcript in uninfected plant tissue (Fig. 1B).
Figure 1.
Analysis of Bs3 and Bs3-E transcript abundance. Transcript abundance was determined by semiquantitative RT-PCR. The constitutively expressed gene elongation factor 1α (EF1α) served as a normalization control. A, An increase of Bs3 transcript abundance was detectable 6 h post infection (hpi) with Xcv strain 85-10 expressing avrBs3. Leaves of 5- to 6-week-old plants of pepper cv ECW-30R (Bs3 genotype) were inoculated with Xcv (OD600 = 0.4) via blunt syringe. Inoculated leaf tissue was harvested at 0, 3, 6, 9, 12, 18, and 24 h post infection, and RNA was extracted and reverse transcribed into cDNA. To determine the earliest time point at which Bs3 transcript was detectable, 35 and 40 PCR cycles were carried out. B, Bs3 and Bs3-E transcripts were detectable only after infection with Xcv strains expressing the matching Avr protein. Tissue-specific analysis of Bs3 and Bs3-E transcripts was performed on cDNA from uninfected leaf, flower, fruit, and root tissue of pepper ECW-30R (Bs3) and ECW (Bs3-E). As a positive control for RT-PCR, we used cDNA derived from ECW-30R and ECW leaves that were inoculated with avrBs3- and avrBs3Δrep16-expressing Xcv strains (OD600 = 0.4), respectively.
To define the minimal Bs3 and Bs3-E promoter regions, we generated progressive 5′ deletions and fused these to the Bs3 and Bs3-E coding sequences (cds). Functionality and specificity of corresponding T-DNA constructs were tested in leaves of Nicotiana benthamiana by Agrobacterium tumefaciens-mediated codelivery of a cauliflower mosaic virus 35S (35S) promoter-driven avrBs3 (35S:avrBs3) or avrBs3Δrep16 (35S:avrBs3Δrep16) gene, respectively (Fig. 2). Functional Bs3 and Bs3-E promoter deletion derivatives were expected to be transcriptionally induced by AvrBs3 and AvrBs3Δrep16, respectively, resulting in the expression of the Bs3 and Bs3-E proteins and triggering an HR.
Figure 2.
The UPA boxes of the Bs3 and Bs3-E promoters are crucial to their inducibility by the matching AvrBs3 and AvrBs3Δrep16 proteins. To define the minimal Bs3 and Bs3-E promoters, progressive 5′ deletions of the Bs3 promoter (343 Bs3, 166 Bs3, and 90 Bs3; A and B) and the Bs3-E promoter (356 Bs3-E, 179 Bs3-E, and 90 Bs3-E; C and D) were fused to the Bs3 and Bs3-E cds, respectively. The Bs3 and Bs3-E promoter constructs were delivered as denoted into N. benthamiana leaves via A. tumefaciens (OD600 = 0.8) together with constructs containing the 35S promoter-driven avrBs3Δrep16 (left side of the leaf; + avrBs3Δrep16) or avrBs3 (right side of the leaf; + avrBs3) genes (B and D). In addition, avrBs3 and avrBs3Δrep16 were expressed individually in the absence of a Bs3 or a Bs3-E promoter construct (avrBs3* or avrBs3Δrep16*). Dashed lines mark inoculated areas. Four days post infiltration (dpi), leaves were harvested and cleared with ethanol to visualize the HR (dark areas in B and D). Schemes in A and C show the length of the Bs3 promoter (A) and the Bs3-E promoter (C) deletions examined. Promoter deletion constructs are designated according to the length of the respective promoter fragment with respect to the ATG start codon and are displayed to scale. The total length of promoter regions and the 5′ UTR of Bs3 and Bs3-E are 1,023 and 1,036 bp, respectively. All similarly sized Bs3 and Bs3-E promoters are identical in their 5′ and 3′ ends but differ in size due to the presence of a 13-bp insertion/deletion polymorphism that is present in the Bs3-E promoter and absent from the Bs3 promoter. Hatched boxes represent the promoter and the 5′ UTR of the genes. Small white and black boxes represent the UPA boxes from the Bs3 (UPAAvrBs3) and Bs3-E (UPAAvrBs3Δrep16) promoters, respectively. Note that the Bs3 and Bs3-E promoters differ only within these UPA boxes but are otherwise identical and therefore are displayed in identical color. Gray boxes represent the cds of the Bs3 and Bs3-E genes. + and – indicate the presence and absence of an HR in N. benthamiana (A and C).
Data observed with overexpression of bacterial effectors in a heterologous system generally need to be treated with caution. Yet, we have previously demonstrated that Bs3-mediated recognition specificity is unaffected even if highly related TALes are expressed to high levels in a heterologous system (Schornack et al., 2005; Römer et al., 2007). Furthermore, our assay reflects the natural situation where activation of the Bs3 promoter results in the HR (Römer et al., 2007).
Using A. tumefaciens-mediated delivery, it was found that Bs3 promoter fragments containing 1,023, 343, or 166 bp upstream of the ATG start codon triggered the HR in combination with 35S:avrBs3 but not with 35S:avrBs3Δrep16 (Fig. 2B). By contrast, a 90-bp Bs3 promoter fragment, which lacks the previously predicted UPAAvrBs3 box (TATATAAACCN2-3CC; Kay et al., 2007), was not able to trigger the HR in N. benthamiana when codelivered with the 35S:avrBs3 construct. In addition, we tested a set of equivalent Bs3-E promoter deletion derivatives (Fig. 2C; note that similarly sized Bs3 and Bs3-E promoter deletions are identical at their 5′ and 3′ termini but differ in size due to a 13-bp insertion/deletion polymorphism). As observed with the Bs3 promoter deletion derivatives, a Bs3-E promoter deletion that contains only 90 bp upstream of the ATG start codon (Fig. 2D; construct 90 Bs3-E) did not trigger HR when codelivered with a 35S:avrBs3Δrep16 construct. The exact location of the UPAAvrBs3Δrep16 box of the Bs3-E promoter has not been defined; however, it likely will include the 13-bp insertion/deletion polymorphism present in the Bs3-E promoter and absent from the Bs3 promoter, because this is the only difference between the two promoters. Consistent with this region playing a role in activation is the result that the uninducible promoter derivatives 90 Bs3 and 90 Bs3-E do not contain the regions that are polymorphic between Bs3 and Bs3-E.
To determine if other sequences, apart from the predicted UPAAvrBs3 box, are crucial to AvrBs3-mediated transcriptional activation, we generated a set of Bs3 promoter mutants carrying different deletions between the ATG start codon and the UPAAvrBs3 box (Fig. 3A). The Bs3 promoter deletions were fused in front of the Bs3 cds and were delivered into N. benthamiana leaves via A. tumefaciens T-DNA transfer together with a 35S:avrBs3 gene (Fig. 3B). A Bs3 promoter deletion derivative that lacks a 44-bp region 3′ of the UPAAvrBs3 box (Bs3Δ56-99; Fig. 3A) was still capable of triggering an AvrBs3-dependent HR. By contrast, Bs3 promoter mutants with deletions larger than 59 bp were not functional. These data suggest that a minimum distance between the UPAAvrBs3 box and the ATG start codon is needed in order to generate a transcript that encodes a functional Bs3 protein. We carried out RACE to determine the transcriptional start site (TSS) of all constructs. As anticipated, most constructs that did not mediate an AvrBs3-dependent HR produced transcripts that lack the ATG start codon of the Bs3 cds (constructs Bs3Δ1-99, Bs3Δ1-91, and Bs3Δ1-80; Fig. 3A; Supplemental Data Set S1). Construct Bs3Δ1-59, which also does not mediate an AvrBs3-dependent HR, produces a transcript that starts with the ATG start codon of the Bs3 cds but that contains no 5′ untranslated region (UTR). It seems likely that the absence of an HR with construct Bs3Δ1-59 is due to the lack of a 5′ UTR. Therefore, an essential aspect in the analysis of UPA box-containing promoters is to consider a spacing between the UPA box and the start of transcription.
Figure 3.
Extended Bs3 promoter deletions 3′ of the UPAAvrBs3 box suppress the AvrBs3-triggered and Bs3-mediated HR. To analyze if sequences apart from the UPAAvrBs3 box are needed for AvrBs3-mediated activation of the Bs3 promoter, regions between the UPAAvrBs3 box and the ATG start codon were deleted. A, Graphical display of Bs3 promoter deletion constructs. The designation of the deletions refers to the first and last deleted bp in the given promoter starting from the first bp 5′ of the ATG start codon. Deletions are displayed as white boxes, and their size is indicated in bp (triangles). The Bs3 promoter region, the UPAAvrBs3 box, and the Bs3 cds are displayed as hatched, black, and gray boxes, respectively. The indicated 5′ UTR refers to the Bs3 wild-type gene. Arrows above the boxes mark the TSS of the given promoter. The scale at the bottom indicates the distances with respect to the ATG start codon. With the exception of the Bs3 cds, all elements are drawn to scale. All deletions were fused individually in front of the Bs3 cds. + and – indicate the presence and absence of an HR in N. benthamiana for each construct when being codelivered with a 35S:avrBs3 T-DNA. The sequences, deletions, and TSS of all promoters are also provided as Supplemental Data Set S1. B, Functional analysis of Bs3 promoter deletion constructs. Individual deletion constructs were delivered together with 35S:avrBs3 via A. tumefaciens (OD600 = 0.8) into N. benthamiana leaves. Dashed lines mark the inoculated areas. The leaves were harvested at 4 dpi and cleared with ethanol to visualize the HR (dark areas).
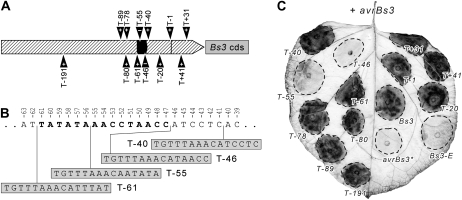
While a deletion analysis is capable of coarse characterization of regulatory regions in a promoter, linker-scanning mutagenesis permits a higher resolution identification of short, defined sequence motifs and their effect on promoter activity (McKnight and Kingsbury, 1982). We analyzed a set of 12 linker-scanning Bs3 promoter mutants. Each mutant contained a 15-bp insertion located between positions +31 and −191 relative to the TSS (Fig. 4, A and B; Supplemental Data Set S2). The Bs3 promoter mutants were fused to the Bs3 cds and were delivered via A. tumefaciens T-DNA transfer together with the 35S:avrBs3 construct into N. benthamiana leaves (Fig. 4C) to test for HR induction. Two of the 12 mutants (T−46 and T−55) no longer triggered an HR. The insertions that affected Bs3 promoter function are within (T−55) or adjacent to (T−46) the previously defined UPAAvrBs3 box (Fig. 4B) that spans a region from −47 to −61 bp. Given that the T−46 insertion is located adjacent to but not within the previously predicted UPAAvrBs3 box, the functionally relevant nucleotides of the box likely extend farther into the 3′ region of the promoter. Given that all insertions that affected Bs3 promoter function were located at or in the UPAAvrBs3 box, it seems likely that this is the only sequence motif that is crucial to AvrBs3-mediated promoter activation.
Figure 4.
Linker-scanning mutagenesis of the Bs3 promoter region identifies the UPAAvrBs3 box as a functionally crucial element. A, Distribution of transposon footprints in the Bs3 promoter. To identify sequences that are crucial for AvrBs3-mediated transcriptional activation of the Bs3 promoter, linker-scanning mutagenesis was carried out. Twelve transposon footprint mutants (T; black triangles above or below the hatched box) were obtained, containing a 15-bp insertion. Numbering of the mutants refers to the nucleotide that is adjacent to the 3′ end of the respective transposon footprint with respect to the TSS. The Bs3 promoter region and the 5′ UTR are displayed as hatched areas. The transcriptional start site of the wild-type Bs3 gene is indicated with a black horizontal line. The UPAAvrBs3 box and the Bs3 cds are displayed as black and gray boxes, respectively. + and – indicate the presence and absence of an HR with each promoter construct in N. benthamiana leaves when codelivered with a 35S:avrBs3 T-DNA (+ avrBs3). avrBs3* denotes a tissue patch in which no Bs3 promoter construct but only a 35S:avrBs3 T-DNA was delivered. The 5′ UTR in the Bs3 wild-type promoter is 59 bp in size, and thus insertions T + 31 and T + 41 are located in the 5′ UTR. B, Insertions adjacent to or within the predicted UPAAvrBs3 box. The numbers above the nucleotide sequence indicate the distance with respect to the TSS. Boldface letters represent the predicted UPAAvrBs3 box. Sequences of transposon mutants are available as Supplemental Data Set S2. C, Functional analysis of linker-scanning-derived Bs3 promoter mutants. Each mutant was fused in front of the Bs3 cds and was delivered together with 35S:avrBs3 into N. benthamiana leaves via A. tumefaciens (OD600 = 0.8). Dashed lines mark the inoculated areas. Leaves were harvested at 4 dpi and cleared with ethanol to visualize the HR (dark areas).
Recognition Specificity of the Bs3-E Promoter
The Bs3 and Bs3-E promoters differ only by a 13-bp insertion (CTCTATTCCACTA) in Bs3-E compared with Bs3 (Römer et al., 2007). Although it is conceivable that this polymorphic area defines recognition specificity, the particular sequences that contribute to specificity in the Bs3-E promoter remained unclear. We hypothesized that the 13-bp insertion in the Bs3-E promoter contains the complete UPAAvrBs3Δrep16 box of the Bs3-E promoter. Here, we tested this hypothesis by placing the 13-bp insertion of the Bs3-E promoter into two different locations of the Bs3 promoter and fused the Bs3 promoter derivatives (Bs3 −20i and Bs3 +31i) in front of the Bs3 cds (Fig. 5A). We anticipated that Bs3 −20i and Bs3 +31i would be activated by both AvrBs3 and AvrBs3Δrep16. However, our data demonstrated that only 35S:avrBs3 triggered the HR in combination with Bs3 −20i and Bs3 +31i, whereas 35S:avrBs3Δrep16 failed to trigger the HR with these promoter derivatives (Fig. 5B). Based on these data, it appears that the 13-bp insertion represents a part of, but not the complete, UPAAvrBs3Δrep16 box.
Figure 5.
The Bs3-E promoter-specific 13-bp insertion exerts its function on recognition specificity in a position-dependent manner. A, The 13-bp insertion sequence (CTCTATTCCACTA) that is specific to the Bs3-E promoter (insertion at position −50 of the Bs3 promoter) was placed into the promoter and 5′ UTR of the Bs3 gene. Numbering of the corresponding mutant derivatives (Bs3 −20i and Bs3 +31i) refers to the nucleotide in the Bs3 promoter that is adjacent to the 3′ end of the 13-bp insertion. The nucleotide sequence highlighted in boldface letters represents the UPAAvrBs3Δrep16 box (see Fig. 7). The Bs3 promoter region and the 5′ UTR are displayed as a hatched box. The UPAAvrBs3 box and the Bs3 cds are displayed as black and gray boxes, respectively. B, Functional analysis of the Bs3 promoter derivatives Bs3 −20i and Bs3 +31i. Both Bs3 promoter derivatives were fused to the Bs3 cds and were codelivered with 35S promoter-driven avrBs3 (+ avrBs3) or avrBs3Δrep16 (+ avrBs3Δrep16) genes into N. benthamiana leaves via A. tumefaciens (OD600 = 0.8). Leaf areas in which only a 35S:avrBs3 or a 35S:avrBs3Δrep16 T-DNA was delivered are marked as avrBs3* or avrBs3Δrep16*, respectively. Dashed lines mark the inoculated areas. Leaves were harvested at 4 dpi and cleared with ethanol to visualize the HR (dark areas).
To further define the recognition specificity of the Bs3-E promoter, we placed insertions of one, two, or three nucleotides into the Bs3 promoter instead of the 13-bp sequence (Fig. 6A). The Bs3 promoter insertion mutants were fused to the Bs3 cds and delivered via A. tumefaciens together with 35S:avrBs3Δrep16 or 35S:avrBs3 into N. benthamiana leaves. We found that only the insertions Bs3+CT, Bs3+CTA, and Bs3+CTC triggered the HR in combination with AvrBs3Δrep16 (Fig. 6). By contrast, all other insertions of one, two, and three nucleotides did not result in an AvrBs3Δrep16-responsive promoter (Fig. 6B). We concluded that the CTC motif at the 5′ end of the 13-bp insertion in the Bs3-E promoter is part of the UPAAvrBs3Δrep16 box. Given that not only a CTC but also a CTA insertion triggered the AvrBs3Δrep16-dependent HR, it seems likely that the C nucleotide at the 3′ terminal end can be functionally replaced by an A nucleotide. Since the Bs3+CT insertion is followed by an A nucleotide, this promoter mutant also contains a CTA motif at the 5′ end of the insertion site. This probably explains why Bs3+CT is functionally identical to the Bs3+CTA construct.
Figure 6.
An insertion of 2 bp into the Bs3 promoter causes a change in recognition specificity. A, Schematic representation of insertions that were placed into the Bs3 promoter region. All insertions are indicated below the Bs3 promoter with yellow background. The 13-bp natural insertion of the Bs3-E promoter is displayed above the Bs3 promoter with yellow background. Insertions were placed into the Bs3 promoter at the position where the Bs3-E promoter contains the 13-bp insertion with respect to the Bs3 promoter. Red letters highlight insertions corresponding to the 5′ end of the 13-bp insertion in the Bs3-E promoter. Brown and blue letters display the predicted UPAAvrBs3 box of the Bs3 promoter and the experimentally defined UPAAvrBs3Δrep16 box of the Bs3-E promoter, respectively (see Fig. 7). Green and gray boxes represent the Bs3 promoter and the Bs3 cds, respectively. + and – indicate the presence and absence of an HR in N. benthamiana leaves upon codelivery of each construct with a 35S promoter-driven avrBs3 (AvrBs3) or avrBs3Δrep16 (AvrBs3Δrep16) gene, respectively. B, Functional analysis of Bs3 promoter insertion mutants. Representative mutants were expressed transiently in N. benthamiana leaves via A. tumefaciens (OD600 = 0.8) together with T-DNA constructs containing 35S promoter-driven avrBs3 (+ avrBs3) or avrBs3Δrep16 (+ avrBs3Δrep16) genes. Leaf areas in which only a 35S:avrBs3 or a 35S:avrBs3Δrep16 T-DNA was delivered are marked as avrBs3* or avrBs3Δrep16*, respectively. Dashed lines mark the inoculated areas. Leaves were harvested at 4 dpi and cleared with ethanol to visualize the HR (dark areas).
Systematic Substitution Mutagenesis of the Bs3 Promoter
Previously, the sequence of the UPAAvrBs3 box was determined to be TATATAAACCN2-3CC by comparing three different AvrBs3-inducible promoters (Kay et al., 2007; Römer et al., 2007). However, in this study, we found that the Bs3 promoter mutant T−46, which contains an insertion adjacent to the 3′ end of the predicted UPAAvrBs3 box, was no longer capable of triggering an AvrBs3-dependent HR (Fig. 4). This suggested that the UPAAvrBs3 box extends farther into the 3′ direction than previously assumed. To determine the functionally relevant nucleotides of the UPAAvrBs3 box more precisely, we performed a systematic substitution mutagenesis of the Bs3 promoter to permutate each base from position −41 to −63 (Fig. 7A). The resulting 69 Bs3 promoter substitution mutants were fused in front of the Bs3 cds and were delivered via A. tumefaciens together with 35S:avrBs3Δrep16 or 35S:avrBs3 into N. benthamiana leaves. We identified three functionally distinct classes of substitution mutants. Forty-two substitution mutants were functionally identical to the Bs3 wild-type promoter and triggered an HR in combination with 35S:avrBs3 but not in combination with 35S:avrBs3Δrep16 (Fig. 7B, light green boxes). Twenty substitution mutants produced no HR in combination with either 35S:avrBs3 or 35S:avrBs3Δrep16 (Fig. 7B, red boxes). The remaining seven substitution mutants showed a reduced HR phenotype (Fig. 7B, boxes with red and light green triangles). This reduced HR phenotype was observed in multiple repetitions and thus is highly reproducible. Inspection of the Bs3 promoter substitution mutants revealed that functionally relevant nucleotides of the UPAAvrBs3 box span the region from −61 to −44. Thus, the experimentally defined UPAAvrBs3 box (TATATAAACCTAACCATC) extends three nucleotides farther in the 3′ direction as compared with the UPAAvrBs3 box described previously (TATATAAACCN2-3CC; Fig. 7; Kay et al., 2007). Based on the experimentally defined UPAAvrBs3 box, the Bs3 promoter mutant T−46 (Fig. 4B) contains an insertion within and not adjacent to the UPAAvrBs3 box and explains why this insertion mutant is incapable of triggering the AvrBs3-dependent HR (Fig. 4C). Another notable observation of our systematic mutagenesis is that the functional consequences of substitutions in the UPAAvrBs3 box differ depending on which position is mutated. For example, any mutation at position −61, −57, or −47 abolished the AvrBs3-mediated HR (Fig. 7B). By contrast, none of the substitutions at positions −59, −50, −49, −46, and −45 had a detectable effect on the AvrBs3-mediated HR.
Figure 7.
Substitution mutagenesis of the Bs3 and Bs3-E promoters permits exact containment of the corresponding UPA boxes. A, Generation of Bs3 and Bs3-E promoter substitution mutants. Individual nucleotides in the Bs3 or Bs3-E promoter were replaced by all three alternative nucleotides via site-directed mutagenesis. The top sequence is part of the Bs3 wild-type promoter. Nucleotide positions are relative to the TSS of the Bs3 promoter. The five sequences below refer to representative substitution mutants. Mutagenized nucleotides are displayed in red letters. In total, 69 Bs3 and 30 Bs3-E substitution mutants were generated. B, Functional analysis of Bs3 promoter mutants. The 69 different Bs3 promoter mutants and the wild-type Bs3 promoter were fused to the Bs3 cds and were codelivered with a 35S promoter-driven avrBs3 construct into N. benthamiana leaves via A. tumefaciens (OD600 = 0.8). The phenotypes were scored at 4 dpi. The colored boxes summarize the results of the phenotypic scoring. Nucleotide positions displayed in the top row are numbered relative to the TSS of Bs3. Green boxes display the wild-type Bs3 promoter that triggers the HR. Each light green box represents a substitution mutant that triggers the HR. Substitution mutants that did not trigger the HR are displayed in red. Boxes representing substitution mutants with an intermediate phenotype (weak HR) are displayed as light green and red triangles. Please note that the collection of green boxes represents one data point (the wild-type Bs3 promoter), while the other boxes represent the results that were observed with distinct substitution mutants. The UPAAvrBs3 box deduced from this analysis is shown at bottom in blue letters. The underlined sequence represents the previously predicted UPAAvrBs3 box (Kay et al., 2007). C, Functional analysis of Bs3-E promoter mutants. Thirty distinct Bs3-E promoter mutants and the wild-type Bs3-E promoter were fused to the Bs3-E cds and delivered with a 35S:avrBs3Δrep16 construct into N. benthamiana leaves via A. tumefaciens (OD600 = 0.8). The phenotypes were scored at 4 dpi. Color coding is as in B, with the difference that green boxes represent the Bs3-E promoter. Nucleotide positions that are displayed in the top row of boxes are relative to the TSS of Bs3-E. The deduced minimal UPAAvrBs3Δrep16 box of Bs3-E is displayed in brown letters. Yellow background marks the Bs3-E promoter-specific 13-bp insertion. The gray background marks corresponding positions in the Bs3 and Bs3-E promoters, which yielded almost identical results in the substitution mutant analysis.
Substitution Mutagenesis of the Bs3-E Promoter
To analyze the UPAAvrBs3Δrep16 box, we carried out substitution mutagenesis of selected positions within the Bs3-E promoter (Fig. 7C). The observation that Bs3 promoter mutants containing a 3-bp CTC insertion triggered an HR in combination with AvrBs3Δrep16 (Fig. 6) suggests that this sequence motif defines the 3′ end of the UPAAvrBs3Δrep16 box. In agreement with this hypothesis, we identified Bs3-E promoter substitution mutants in the CTC motif (positions −63 to −61) that do not mediate the HR in combination with AvrBs3Δrep16 (Fig. 7C). All Bs3-E promoter mutants that contained substitutions located in the 3′ direction of position −61 were functionally identical to the Bs3-E wild-type promoter. By contrast, many Bs3-E promoter mutants that contained substitutions located in the 5′ direction of the CTC motif lost the capability to mediate an AvrBs3Δrep16-triggered HR (Fig. 7C). Taken together, our data define the UPAAvrBs3Δrep16 box region as extending from position −74 to position −61 (TATATAAACCTCTC; Fig. 7C). Although it remains to be clarified if the UPAAvrBs3Δrep16 box extends at its 5′ end beyond position −74, it seems evident that the UPAAvrBs3 and the UPAAvrBs3Δrep16 boxes overlap in at least 11 nucleotides (Fig. 7C). In this context, it is notable that substitution mutations in corresponding positions in the UPAAvrBs3 and UPAAvrBs3Δrep16 boxes (−61 [Bs3] versus −74 [Bs3-E], −57 [Bs3] versus −70 [Bs3-E], and −53 [Bs3] versus −66 [Bs3-E]) have almost identical functional consequences. For example, all substitutions in position −57 of the Bs3 promoter and in position −70 of the Bs3-E promoter resulted in nonfunctional promoters (Fig. 7, B and C). Similarly, substitutions to an A nucleotide at position −53 of the Bs3 and position −66 of the Bs3-E promoter had no obvious functional consequences, while mutations to G or T resulted in nonfunctional Bs3 or Bs3-E promoter mutants.
DNA-Binding Specificity of AvrBs3 and AvrBs3Δrep16
Previously, EMSA showed that AvrBs3 binds with high affinity to the Bs3 promoter and weakly to the Bs3-E promoter (Römer et al., 2007). Unexpectedly, EMSA showed also that AvrBs3Δrep16 had a higher affinity to the Bs3 promoter as compared with the Bs3-E promoter fragment (Römer et al., 2007). EMSA with AvrBs3Δrep16 and AvrBs3 was carried out with DNA probes of identical size that had identical sequence at their 3′ end but not at their 5′ end (Fig. 8A). Based on our data described above, it is now clear that the Bs3-E promoter probe did not span the complete UPAAvrBs3Δrep16 box, which likely accounts for the unexpected EMSA results (Römer et al., 2007). Therefore, we repeated the EMSA with Bs3 and Bs3-E promoter-derived probes that are sequence identical at their 3′ and 5′ ends but that, due to the 13-bp insertion in the Bs3-E promoter, are not identical in size. Hence, the new probe for the Bs3-E promoter is likely to contain the complete UPAAvrBs3Δrep16 box. As shown in Figure 8, GST:AvrBs3Δrep16 bound with high affinity to the Bs3-E-derived and with low affinity to Bs3-derived promoter fragments (Fig. 8, B and C). Similarly, GST:AvrBs3 binds with high and low affinity to the Bs3- and Bs3-E-derived promoter fragments, respectively. Competition assays with labeled Bs3-derived promoter fragments and nonlabeled Bs3- and Bs3-E-derived promoter fragments, and vice versa, further confirm that AvrBs3 has high affinity to the Bs3 promoter fragment and only low affinity to the Bs3-E promoter fragment (Fig. 8D). Competition assays for AvrBs3Δrep16 showed that it binds with high affinity to the Bs3-E promoter fragment and with low affinity to the Bs3 promoter fragment. Together, these data strongly suggest that, in contrast to our previous statements (Römer et al., 2007), specific binding of AvrBs3 or AvrBs3Δrep16 to their matching promoters is the basis for promoter activation specificity.
Figure 8.
AvrBs3 and AvrBs3Δrep16 bind with high affinity to the Bs3 and Bs3-E promoters, respectively. A, Probes derived from the Bs3 and Bs3-E promoter sequences used in EMSA. The experimentally defined UPAAvrBs3 and UPAAvrBs3Δrep16 boxes of the Bs3 and Bs3-E promoters (see Fig. 7) are displayed as boldface black and gray letters, respectively. Numbering is relative to the TSS of Bs3 and Bs3-E, respectively. Hatched background indicates the 13-bp insertion of the Bs3-E promoter. Sequences of biotin-labeled probes are indicated by lines above and below the promoter sequences. Black and gray lines mark the Bs3 and Bs3-E promoter probe sequences used in these experiments; the dashed line marks the Bs3-E promoter probe sequence used previously (Römer et al., 2007). Note that the probe sequence used previously (dashed line) does not cover the entire UPAAvrBs3Δrep16 box (gray letters). B, EMSA of 100 fmol of biotin-labeled Bs3-derived (36 bp) and Bs3-E-derived (49 bp) probes incubated with 100, 250, and 500 fmol of GST:AvrBs3 (AvrBs3) and GST:AvrBs3Δrep16 (AvrBs3Δrep16) fusion proteins, respectively. Positions of bound and free probes are indicated: arrow, bound probe; asterisk, free probe. The top signals correspond to the slots. C, Coomassie Brilliant Blue-stained 8% SDS-PAGE. GST translational fusions to AvrBs3 and AvrBs3Δrep16 used for EMSA studies were expressed in Escherichia coli, purified, and quantified by Bradford analysis (Bradford, 1976). Subsequently, 1.5 and 3 μg of GST:AvrBs3, GST:AvrBs3Δrep16, and bovine serum albumin (BSA) standard were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. Fragments of the expected size (GST:AvrBs3, 150.8 kD; AvrBs3Δrep16, 136.8 kD) are indicated by asterisks. Marker proteins (M) are indicated with their molecular masses in kD (PageRuler prestained protein ladder; Fermentas). D, EMSA competition assay of 100 fmol of biotin-labeled Bs3 (left) or Bs3-E (right) probe incubated with 500 fmol of GST:AvrBs3 (AvrBs3) or GST:AvrBs3Δrep16 (AvrBs3Δrep16), respectively. A molar excess of nonlabeled Bs3 and Bs3-E fragments of 25×, 50×, and 100× was used for competition. All experiments were repeated twice with similar results.
A Bs3 Promoter with an Inverted UPAAvrBs3 Box Does Not Trigger the Bs3-Mediated HR
To clarify if the UPAAvrBs3 box acts in a directional manner, we generated a Bs3 promoter derivative in which the UPAAvrBs3 box is replaced by the reverse-complement sequence (Fig. 9A). The corresponding Bs3 promoter mutant (Bs3 UPArev) was fused to the Bs3 cds and was delivered via A. tumefaciens together with 35S:avrBs3 into N. benthamiana leaves. The Bs3 UPArev construct did not trigger the HR in combination with AvrBs3, suggesting that the UPAAvrBs3 box acts in a directional manner (Fig. 9B).
Figure 9.
A Bs3 promoter with an inversely orientated UPAAvrBs3 box does not trigger a Bs3-dependent HR. A, Part of the Bs3 promoter wild-type sequence (Bs3) and a derivative with an inverted UPAAvrBs3 box (Bs3 UPArev) are shown, highlighted with gray boxes. The experimentally determined UPAAvrBs3 box is shown in white letters (see Fig. 7). Numbering is relative to the TSS of Bs3. B, Functional analysis of the inverted box in the Bs3 promoter. The promoters depicted in A were fused to the Bs3 cds and delivered via A. tumefaciens (OD600 = 0.8) alone (asterisks) or together with a 35S promoter-driven avrBs3 gene (+ avrBs3) into N. benthamiana leaves. Dashed lines mark the inoculated areas. Leaves were harvested at 4 dpi and cleared with ethanol to visualize the HR (dark areas).
Functionality of the UPAAvrBs3 Box Is Independent of the Promoter Context
The 3′ end of the UPAAvrBs3 box in the Bs3 promoter is located 44 bp upstream of the TSS and 102 bp upstream of the ATG start codon. We wondered if the UPAAvrBs3 box retains its functionality when moved to other positions within the Bs3 promoter. As starting material for this experiment, we used a Bs3 promoter substitution mutant (designated Bs3 UPAmut; the wild-type T nucleotide at position −61 is replaced by an A nucleotide) that, when fused in front of the Bs3 cds, did not trigger the HR in N. benthamiana leaves upon A. tumefaciens-mediated codelivery with 35S:avrBs3 (Fig. 10). Next, we inserted a wild-type UPAAvrBs3 box into Bs3 UPAmut at a distance of 293 bp (Bs3 UPA293) or 424 bp (Bs3 UPA424) upstream of the ATG start codon. Bs3 UPA293 and Bs3 UPA424 promoter constructs were fused in front of the Bs3 cds and were tested functionally via A. tumefaciens-mediated delivery. Both promoter constructs triggered an HR in N. benthamiana upon codelivery with 35S:avrBs3 (Fig. 10B). Thus, the UPAAvrBs3 box retains its functionality when moved within the Bs3 promoter. Next, we tested if the UPAAvrBs3 box would retain its function when moved to a promoter context different from the pepper Bs3 promoter. We used the promoter of the tomato R gene Bs4, which is transcribed constitutively at a low level (Schornack et al., 2005) and which does not trigger the HR when fused in front of the Bs3 cds (Fig. 10B). We placed the UPAAvrBs3 box and a mutated box variant into the Bs4 promoter (termed Bs4 UPA and Bs4 UPAmut, respectively) and fused these Bs4 promoter derivatives in front of the Bs3 cds. A. tumefaciens-mediated delivery of 35S:avrBs3 triggered an HR in N. benthamiana leaves upon codelivery of the Bs4 UPA but not the Bs4 UPAmut promoter construct. Thus, the UPAAvrBs3 box retains its functionality not only in different locations within the pepper Bs3 but also in the context of the tomato Bs4 promoter.
Figure 10.
Functionality of the UPAAvrBs3 box is independent of its position and the promoter context. A, Graphical representation of promoter constructs tested. Mutations and insertions of UPAAvrBs3 boxes were made via site-directed mutagenesis. Green boxes represent the Bs3 and Bs3-E promoters. Yellow boxes represent the Bs4 promoter. Blue and brown boxes represent the UPAAvrBs3 and UPAAvrBs3Δrep16 boxes, respectively. Note that the Bs3 and Bs3-E promoters differ only within and adjacent to their UPA boxes but are otherwise identical and therefore displayed in identical color. A white vertical line within the UPAAvrBs3 box represents a substitution mutation (T→A at position −61; see Fig. 7B) within the UPAAvrBs3 box. All promoter elements are displayed to scale. The numbers and horizontal lines above the promoters provide a scale and denote the distances with respect to the ATG start codon. Arrows above the promoter boxes mark the TSS of the given promoter. Numbers below the arrows denote the distance between the 3′ end of the UPAAvrBs3 box and the respective TSS. Gray boxes represent the Bs3 cds. + and – indicate the presence and absence of an HR in N. benthamiana upon codelivery of each promoter construct with a 35S:avrBs3 construct. B, Functional analysis of Bs3 and Bs4 promoter derivatives. The promoter constructs depicted in A were codelivered together with a 35S:avrBs3 construct (+ avrBs3) into N. benthamiana leaves via A. tumefaciens (OD600 = 0.8). Dashed lines mark the inoculated areas. Four days after inoculation, the leaves were harvested and cleared with ethanol to better visualize the HR (dark areas).
The TSS of the Bs3 promoter is 44 bp downstream of the 3′ end of the UPAAvrBs3 box (Römer et al., 2007). We carried out RACE to determine the TSS generated in constructs that trigger the HR in combination with 35S:avrBs3 (Bs3 UPA293, Bs3 UPA424, and Bs4 UPA). As shown in Figure 10A, the TSS of Bs3 UPA293, Bs3 UPA424, and Bs4 UPA were 46, 41, and 42 bp downstream of the 3′ end of the respective UPAAvrBs3 boxes. Hence, the TSS in AvrBs3-inducible promoters appears to be dictated by the location of the UPAAvrBs3 box.
Naturally Occurring Nonfunctional Bs3 Alleles
To study natural diversity of Bs3 alleles, we examined accessions of the Capsicum species C. annuum, C. baccatum, and C. chinense. We determined the promoter sequences (244 bp 5′ of the ATG start codon) and cds of 49 Capsicum accessions, identifying 23 different haplotypes (Table I; Supplemental Figs. S1 and S2; Supplemental Data Sets S3 and S4). The Bs3-E haplotype (defined as haplotype 1) was the most prevalent, found in nine of 49 sequenced accessions. Within the promoter region, 39 accessions constituting 14 haplotypes were identical to the Bs3-E haplotype. By contrast, we identified only two accessions that were sequence identical to the Bs3 gene in promoter and cds (Table I). We identified two haplotypes (nos. 16 and 23) that carried mutations in the predicted UPAAvrBs3Δrep16 box (Supplemental Fig. S5). Infection experiments showed that neither avrBs3-expressing nor avrBs3Δrep16-expressing Xcv strains triggered the HR in pepper accessions corresponding to haplotypes 16 and 23 (Table I). We PCR amplified and cloned the two corresponding Bs3 alleles (promoter and cds) and tested their functionality via A. tumefaciens-mediated transient expression in N. benthamiana leaves. Both alleles failed to trigger the HR when codelivered with 35S:avrBs3Δrep16. We also cloned the promoters of haplotypes 16 and 23 in front of the Bs3-E cds to test if the lack of functionality of these alleles is due to promoter polymorphisms. Indeed, transcriptional fusions of the Bs3-E cds to the promoters of haplotypes 16 and 23 did not mediate recognition of AvrBs3Δrep16. In a reciprocal experiment, we fused the strong constitutive 35S promoter in front of the cds of haplotypes 16 and 23. Functional analysis demonstrated that the cds of haplotype 23 but not of haplotype 16 triggered an HR. Inspection of the cds of haplotype 16 revealed a G-to-T substitution in the first exon that causes a Gly (GGC)-to-Val (GTC) exchange in a conserved Gly residue of the predicted FAD-binding domain of the Bs3 protein (Table I; Supplemental Figs. S1A and S2). It is likely, therefore, that this difference renders haplotype 16 inactive. In addition to haplotype 16, we found 16 accessions (nine haplotypes [nos. 5, 8–13, 16, and 20]) that, due to polymorphisms in the cds, are unlikely to encode a functional Bs3 protein. One remarkable finding in this context is an 11-bp deletion in exon 1 that is found in 12 accessions (six haplotypes [nos. 8–13]). This deletion results in a frameshift and an early stop codon in the cds (Supplemental Figs. S1A and S2). Similarly, we identified in haplotype 20 a C-to-G substitution in exon 1 that changes a Tyr (TAC) to a stop codon (TAG). We expressed the cds of haplotype 20 and some representative haplotypes that are likely to encode nonfunctional Bs3 proteins under the control of the 35S promoter and confirmed via A. tumefaciens-mediated transient expression that these alleles are indeed incapable of triggering an HR. Altogether, our data show that many Capsicum accessions contain a nonfunctional Bs3 cds. None of these lines with nonfunctional Bs3 alleles showed any observable morphological phenotypes.
Table I.
Functional analysis of naturally occurring Bs3 alleles
| Haplotype No.a | Promoter Type | Promoter, Comments | Coding Sequence, Commentsb | Functional Analysisc
|
|||
|---|---|---|---|---|---|---|---|
| avrBs3d | avrBs3Δrep16e | 35S:cdsf | Prom:Bs3cdsg | ||||
| 1* | Bs3-E | −/− | +/+ | + | + | ||
| 2 | Bs3-E | DNA polymorphisms also present in Bs3 cds | −/n | +/n | |||
| 3‡ | Bs3 | +/+ | −/− | + | − | ||
| 4 | Bs3-E | Cds identical to Bs3 cds | −/− | +/+ | |||
| 5 | Bs3-E | Loss of function due to mutation in second exon (1,093, CCT[P]→CTT[L]) | −/− | −/− | − | ||
| 6 | Bs3-E | −/− | +/+ | ||||
| 7 | Bs3-E | −/− | +/+ | ||||
| 8 | Bs3-E | 11-bp deletion in first exon (Δ595–605) results in a frameshift and thus an early stop codon (haplotypes 8–13) | −/− | −/− | |||
| 9 | Bs3-E | −/− | −/− | − | |||
| 10 | Bs3-E | −/− | −/− | ||||
| 11 | Bs3-E | −/− | −/− | − | |||
| 12 | Bs3-E | −/− | −/− | − | |||
| 13 | Bs3-E | −/− | −/− | ||||
| 14 | Bs3-E | −/− | +/+ | ||||
| 15 | Bs3-E | −/− | +/+ | ||||
| 16 | Unique | C→G (123) substitution in predicted UPAAvrBs3Δrep16 box | Loss of function due to mutation in first exon (366, GGT[G]→GTC[S]) | −/− | −/− | − | − |
| 17 | Bs3-E | n/− | n/+ | + | |||
| 18 | Identical to no. 19 | C→G (161) substitution | n/n | n/n | |||
| 19 | Identical to no. 18 | C→G (161) substitution | −/− | +/+ | |||
| 20 | Identical to no. 22 | C→A (18) substitution | Substitution in first exon (472, TAC[Y]→TAG[stop]) results in a stop codon | −/− | −/− | ||
| 21 | Bs3-E | −/− | −/+ | + | |||
| 22 | Identical to no. 20 | C→A (18) substitution | n/− | n/+ | + | + | |
| 23 | Unique | Deletion (Δ199) and C→T (121) substitution in predicted UPAAvrBs3Δrep16 box | −/− | −/− | + | − | |
Haplotypes 1 and 3 represent the Bs3-E (*) and Bs3 (‡) alleles, respectively. Capsicum accessions corresponding to the haplotypes are provided in Supplemental Figure S1.
Polymorphic nucleotides are underlined, and the encoded amino acids are given in square brackets. Numbering of polymorphisms refers to Supplemental Figures S1 and S2.
Functionality of the Bs3 alleles was analyzed via Xcv inoculation of the pepper accessions and via A. tumefaciens-mediated transient expression in N. benthamiana. Results are displayed in columns
and
, always with the data observed in pepper first and the data observed in N. benthamiana second. The presence or absence of an HR is indicated by + or −, respectively. n, Not tested. Capsicum accessions were tested with Xcv strains expressing either avrBs3 (d) or avrBs3Δrep16 (e). For A. tumefaciens-mediated transient expression in N. benthamiana, the cloned Bs3 alleles (promoter and cds) were delivered in combination with a cauliflower mosaic virus 35S promoter-driven avrBs3 (d) or avrBs3Δrep16 (e) construct.
The cds of the given Bs3 allele was expressed under the control of the 35S promoter (35S:cds).
The promoter of a given Bs3 allele was fused in front of the Bs3 wild-type cds and was delivered into N. benthamiana in combination with a 35S-driven avrBs3Δrep16 T-DNA (Prom:Bs3cds).
DISCUSSION
UPA Boxes Matching to Different TALes Show No Obvious Sequence Homology
We wished to investigate how bacterial TALes specifically interact with and activate corresponding host plant promoters. To do so, we analyzed the Bs3 and Bs3-E promoters to gain insights into how they are specifically activated by the highly related transcription activators AvrBs3 and AvrBs3Δrep16, respectively. Using mutational analysis, we defined the UPAAvrBs3 box of the Bs3 promoter and the UPAAvrBs3Δrep16 box of the Bs3-E promoter (Fig. 7). As both boxes overlap by at least 11 bp, it is unclear if the conserved sequence motif that is present in the UPAAvrBs3 and UPAAvrBs3Δrep16 boxes may also be part of UPA boxes that are targeted by TALes distinct from AvrBs3 and AvrBs3Δrep16. Therefore, we inspected the promoters of the rice (Oryza sativa) genes Xa27, Os8N3, OsTFX1, and OsTFIIAγ1 that have been shown previously to be induced by the matching Xanthomonas oryzae pv oryzae TALes AvrXa27, PthXo1, PthXo6, and PthXo7, respectively (Gu et al., 2005; Chu et al., 2006; Yang et al., 2006; Sugio et al., 2007). None of these promoters contains a sequence that matches the 11-bp sequence present in the UPAAvrBs3 and UPAAvrBs3Δrep16 boxes. Thus, there is no evidence that UPA boxes matching different TALes share a consensus sequence. Our previous studies suggested that the hypervariable residues of TALes determine the promoter target sequence. This hypothesis is based on the analysis of AvrHah1, a TALe from Xanthomonas gardneri, which is recognized in pepper Bs3 plants and thus likely activates the Bs3 promoter. AvrHah1 shares blocks of high homology with AvrBs3 within the hypervariable repeat residues (Supplemental Fig. S3A; Schornack et al., 2008). Thus, comparative analysis of AvrHah1 and AvrBs3 suggested that TALes that share blocks of high homology within their hypervariable repeat residues are likely to target similar UPA boxes. Consistent with this idea is the fact that AvrBs3 and AvrBs3Δrep16 are identical in their first 10 repeat units (Supplemental Fig. S3A) and that the corresponding UPAAvrBs3 and UPAAvrBs3Δrep16 boxes are partially identical. Given that the hypervariable residues of AvrXa27, PthXo1, PthXo6, and PthXo7 share no homology to AvrBs3 within their hypervariable residues (Supplemental Fig. S3, B and C), it is also expected that matching UPA boxes would not share homology with the UPAAvrBs3 box. Taken together, these data indicate that no consensus UPA box exists.
Given that not only the Bs3 but also the pepper UPA10 and UPA20 genes are induced by AvrBs3 (Kay et al., 2007), these promoters should contain a UPAAvrBs3 box. Indeed the UPA10, UPA20, and Bs3 promoters share sequence conservation, which is pronounced in the 5′ end of the corresponding UPAAvrBs3 boxes (positions −61 to −52; Supplemental Fig. S4). Nucleotides at position −46 to −44 of the Bs3 promoter showed less sequence similarity to the corresponding promoter regions of UPA10 and UPA20. In agreement with this observation, our substitution mutagenesis showed that sequence variation at positions −46 and −45 had no detectable effect and substitutions at position −44 had only a minor effect on the Bs3-mediated HR (Fig. 7; Supplemental Fig. S4). In this context, it is notable however, that the Bs3 promoter transposon mutant T−46, which basically lacks the last three nucleotides of the UPAAvrBs3 box (corresponding to positions −46 to −44; Fig. 4), did not trigger an HR. Mutant T−46 is equivalent to a “triple” substitution mutant in which positions −46, −45, and −44 are mutated. Notably, the insertion mutant T−46 showed no Bs3-mediated HR (Fig. 4), while all single nucleotide substitution mutants at positions −46 to −44 showed no or only partial reduction of the Bs3 HR (Fig. 7). We conclude that the suppression of the Bs3 HR in T−46 is due to the additive effect of three substitutions. This implies that single nucleotide substitutions at positions −46 and −45 must affect the inducibility of the Bs3 promoter, although these substitutions had no detectable effect on the Bs3 HR.
Our mutational studies relied on a transient A. tumefaciens-mediated delivery system using constitutively expressed TALe genes in combination with Bs3 and Bs3-E promoter derivatives into N. benthamiana. This assay is useful for rapid analysis of in vitro-generated promoter derivatives. The assay also appears to be highly representative of the native interaction during infection, because naturally occurring Bs3 alleles by Xcv infection assay yielded identical results (see ECW, ECW-30R, PI593576, PI593491, PI631131, PI357635, PI593574, PI406948, PI631152, PI224415, PI599426, PI566809, PI181907, PI497971, CGN17230, CGN17227, PI593557, PI631137, CGN17042, and CGN17025 in Supplemental Fig. S1). Therefore, we are confident that our reporter system is an efficient and accurate assay to study interactions between TALes and corresponding host promoters.
Sequence analysis of naturally occurring Bs3 alleles uncovered that haplotypes 16 and 23 carry mutations in the predicted UPAAvrBs3Δrep16 box (Supplemental Fig. S5). As anticipated, no HR was observed in either haplotype upon infection with avrBs3Δrep16-expressing Xcv strains as well as upon inoculation with an A. tumefaciens strain delivering a 35S promoter-driven avrBs3Δrep16 gene (Table I). Furthermore, when promoters of both haplotypes were fused to the Bs3 wild-type cds, they were unable to trigger an AvrBs3Δrep16-induced HR. Notably, haplotype 16 carries a C-to-G substitution at position −63 relative to the TSS (Supplemental Fig. S5) and thus resembles the C-to-G substitution mutant generated in this study (Fig. 7). By contrast, other haplotypes that contained promoter polymorphisms outside of the UPAAvrBs3Δrep16 box (e.g. haplotypes 19 and 22) were still capable of triggering an AvrBs3Δrep16-dependent HR (Table I). Thus, the definition of the UPAAvrBs3Δrep16 box by the aid of substitution mutants is supported by the functional analysis of naturally occurring Bs3 alleles.
What Defines the TSS in a Gene with a UPA Box?
We discovered that the UPAAvrBs3 box retained its function when moved to different locations within the pepper Bs3 promoter and also in the heterologous tomato Bs4 promoter (Fig. 10). We also determined that the placement of the UPAAvrBs3 box in different promoter locations changed the TSS. The distance between the TSS and the UPAAvrBs3 box was conserved in all constructs and was found to be between 41 and 46 bp with respect to the 3′ end of the UPAAvrBs3 box. Thus, the TSS of AvrBs3-inducible genes seems to be dictated by the location of the UPAAvrBs3 box.
The initiation of mRNA synthesis in eukaryotic cells requires the assembly of general transcription factors and RNA polymerase II into a preinitiation complex at the core promoter. The only known sequence-specific DNA-binding protein among the general transcription factors is the TATA-binding protein, a subunit of the general transcription factor TFIID. Since AvrBs3 was shown to bind to the UPAAvrBs3 box-containing promoters (Kay et al., 2007; Römer et al., 2007) and since the TSS is dictated by the position of the UPAAvrBs3 box, it is possible that AvrBs3 replaces TATA-binding protein in its function as a sequence-specific DNA-binding protein in the preinitiation complex. Given that the UPAAvrBs3 box contains a TATA-like sequence motif, it is tempting to speculate that AvrBs3 binds to this promoter motif. However, mutational analysis of the UPAAvrBs3 box (Fig. 7B) did not provide any evidence that the TATA motif is functionally more important than other areas within the UPAAvrBs3 box.
The observation that the UPAAvrBs3 box works at different promoter locations and within different promoter contexts suggests that diverse UPA boxes corresponding to different TALes may be arranged in tandem to make a complex promoter. If such a complex promoter is fused to the Bs3 cds, it should mediate recognition of multiple TALes. Such a promoter may confer durable resistance against a range of pathogens containing a range of TALes, as is the case of several Xanthomonas species, including X. oryzae pv oryzae, X. oryzae pv oryzicola, and X. axonopodis pv citri.
How Did Bs3 Evolve and Does it Have a Function aside from Resistance?
Previously, we showed that Bs3 and Bs3-E are transcriptionally induced by AvrBs3 and AvrBs3Δrep16, respectively, but not by the TALe AvrBs4 (Römer et al., 2007). Here, we show that Bs3 and Bs3-E transcripts are not detectable via RT-PCR in uninfected leaf, flower, fruit, or root tissue (Fig. 1B). This raises the question of whether the Bs3 protein has a biological function other than in recognition of TALes. Given that expression of Bs3 triggers cell death, a potentially detrimental function that requires strict control of gene expression, one wonders how this gene evolved. The sequence analysis of Bs3 alleles from different Capsicum accessions provides some clues to these questions. About one-third of the analyzed Capsicum accessions (16 of 49) contained a Bs3 cds with a predicted early stop codon (Table I; Supplemental Figs. S1A and S2). It is known that cell death plays an important role not only in disease resistance but also in developmental processes (Lam, 2004). Thus, we wondered if the accessions that carry a nonfunctional Bs3 cds show any phenotypes that would be indicative of a contribution of Bs3 to developmental or physiological processes. Yet, we observed no altered phenotypes in these accessions other than the change in their response to bacterial effectors, indicating that Bs3 is important only in the context of disease resistance.
More than three-quarters of the analyzed Capsicum accessions (38 of 49) were identical to the Bs3-E haplotype within their promoter sequences. About three-quarters of these Bs3-E promoter-containing accessions (29 of 38) differed from the Bs3-E haplotype in their cds. By contrast, the three accessions that contain the Bs3 promoter were identical in their promoter and cds. The Bs3-E haplotype is more prevalent and possibly more ancient than the Bs3 haplotype. Thus, the Bs3 promoter is possibly the consequence of a 13-bp deletion in the Bs3-E promoter. Consistent with this hypothesis, haplotype 4 contains the Bs3-E promoter but is identical to the Bs3 haplotype in its cds (Supplemental Fig. S1A) and thus might represent the progenitor of the Bs3 haplotype.
The predicted Bs3 protein is homologous to YUCCA-like proteins from Arabidopsis (Arabidopsis thaliana), some of which are involved in auxin biosynthesis (Schlaich, 2007; Chandler, 2009). Overexpression of YUCCA-like genes leads to phenotypes characteristic of auxin-overproducing mutants but not to an HR, as in the case of the Bs3 cds (Zhao et al., 2001; Kim et al., 2007; Römer et al., 2007). Hence, YUCCA-like proteins and Bs3 are functionally distinct. Yet, based on sequence homology, Bs3 can be considered as a YUCCA deletion derivative because it lacks a stretch of approximately 70 amino acids present in all predicted YUCCA proteins (Römer et al., 2007). We speculated that the Bs3 gene evolved from a YUCCA-like gene in pepper and anticipated that some Capsicum accessions would contain an ancestral Bs3 gene that encodes a “full-length” YUCCA-like protein instead of the YUCCA deletion derivative. However, none of the Capsicum haplotypes analyzed here contained a Bs3 cds that encodes a full-length YUCCA protein. Thus, the postulated deletion in the Bs3 cds most likely preceded the speciation of Capsicum. Given that Bs3 as a potential YUCCA deletion derivative triggers cell death, its expression must be strictly regulated. Therefore, it appears likely that a promoter mutation that made this promoter transcriptionally inactive preceded the deletion in the cds. In agreement with this idea, knockout mutations in Arabidopsis YUCCA-like genes produced no obvious phenotypes due to genetic redundancy (Schlaich, 2007). In summary, the analyses of Capsicum accessions suggest that the Bs3 gene has no function aside from disease resistance and provide some insights into how and when this potentially detrimental gene evolved.
MATERIALS AND METHODS
Generation of Promoter Deletion Constructs of Bs3 and Bs3-E
Progressive 5′ promoter deletions of the Bs3 gene were PCR amplified from genomic DNA of pepper (Capsicum annuum ‘ECW-30R’; Minsavage et al., 1990). The PCR was carried out with Phusion high-fidelity DNA polymerase (New England Biolabs) and the primers Prom-90 bp-fwd-PR (5′-CACCAGTTATCATCCCCTTTCTCTTTTCTC-3′), Prom-179 bp-fwd-PR (5′-CACCGCACACCCTGGTTAAACAATGAACACG-3′), and Prom-356 bp-fwd-PR (5′-CACCTCATAGTCAAGCTAACGAAACTTATGC-3′) in combination with the primer Final-entry-02-rev (5′-CATTTGTTCTTTCCAAATTTTGGCAATATCTTGTGCAAC-3′). PCR fragments were cloned into pENTR-D (Invitrogen), sequenced, and transferred into the T-DNA vector pGWB1 (Nakagawa et al., 2007) via Gateway recombination (Invitrogen). pGWB1 derivatives were transformed into Agrobacterium tumefaciens GV3101 (Holsters et al., 1980) for transient expression assays. The Bs3-E alleles were cloned in the same way using genomic DNA from pepper cv ECW as template.
Internal promoter deletions were generated by the Phusion site-directed mutagenesis kit (New England Biolabs). We used a Bs3 gene (promoter and cds) cloned in pENTR-D (Invitrogen) as template (Römer et al., 2007) to create deletions. Primers that were used are available upon request. All constructs were sequenced and transferred by Gateway LR recombination into pGWB1 (Nakagawa et al., 2007). pGWB1 derivatives were transformed into A. tumefaciens GV3101 for transient expression assays.
Creation of Promoter Substitution and Insertion Mutants
Substitution mutants in the Bs3 promoter were generated via site-directed mutagenesis using the Phusion site-directed mutagenesis kit (New England Biolabs). We used a Bs3 gene (promoter and cds) cloned in pENTR-D (Invitrogen) as template DNA (Römer et al., 2007). We employed primers that contain at a given position all nucleotides except the nucleotide present in the wild-type sequence. The different permutations were selected by sequence analysis of cloned fragments. The promoter constructs were transferred via Gateway LR recombination into pGWB4 (encodes a C-terminal GFP epitope tag; Nakagawa et al., 2007). pGWB4 derivatives were transformed into A. tumefaciens GV3101 for transient expression in planta. The same approach was used for creation of the Bs3-E promoter mutants, with the difference that we used a cloned Bs3-E gene as template DNA (Römer et al., 2007). For the insertion of nucleotides in the Bs3 promoter, we used the Phusion site-directed mutagenesis kit (New England Biolabs) and the primers site-dir-02-N-fwd-PR (5′-CTGACCAATTTTATTATATAAACCTNAACCATCCTCAC-3′), site-dir-02-NN-fwd-PR (5′-CTGACCAATTTTATTATATAAACCTNNAACCATCCTCAC-3′), site-dir-02-AA+N-fwd-PR (5′-CTGACCAATTTTATTATATAAACCTAANAACCATCCTCAC-3′), or site-dir-02-CT+N-fwd-PR (5′-CTGACCAATTTTATTATATAAACCTCTNAACCATCCTCAC-3′) in combination with the primer site-dir-02-rev-PR (5′-GCAAACGTGTTCATTGTTTAACCAGGGTG-3′). All primers used are phosphorylated at their 5′ termini. We used a Bs3 gene (promoter and cds) cloned in the vector pENTR-D (Invitrogen) as template DNA (Römer et al., 2007). After sequence analysis, cloned fragments were transferred into pGWB1 by Gateway LR recombination (Nakagawa et al., 2007). For insertion of a 13-bp sequence at different locations within the Bs3 promoter, we used the Phusion site-directed mutagenesis kit (New England Biolabs) in combination with primers Prom-Bs3+13-20nU-fwd-PR (5′-CTCTATTCCACTACCTTTCTCTTTTCTCCTCTTG-3′) + Prom-Bs3+13-20nU-rev-PR (5′-GGATGATAACTTGAAGTTGTGGGATG-3′) or primers Prom-Bs3+13+31UTR-fwd-PR (5′-CTCTATTCCACTACAAGTAGTCCTAGTTGCACAT-3′) + Prom-Bs3+13+31UTR-rev-PR (5′-TGTTTTGATAGATTTAGCGGGTGACAAG-3′). A Bs3 gene (promoter and cds) cloned in pENTR-D (Invitrogen) was used as template (Römer et al., 2007). For the insertion of a UPAAvrBs3 box, we also used the Phusion site-directed mutagenesis kit. As template, we used pENTR-D, which contains a Bs3 gene with a mutation (−61T is replaced by A) in the original UPAAvrBs3 box. For the insertion of the Bs3 UPA293 box, we used primers box-02-293-fwd-PR (5′-CAATTTTATTATATAAACCTAACCATCCTCACAACCAAGTAAACTCAAAGAACTAATCATTGAAC-3′) and box-02-293-rev-PR (5′-CATACTAATTTCATATTTCCCTTGCATAAG-3′). To insert the Bs3 UPA424 box, we used primers box-03-424-fwd-PR (5′-CAATTTTATTATATAAACCTAACCATCCTCACAACCACATTAGATTGTACTTGCTTTTTACCACAGATAC-3′) and box-03-424-rev-PR (5′-TCATGTATCATTCGCATTTCAAAGTAAAACTAAGG-3′).
For the generation of Bs4 UPA and Bs4 UPAmut constructs, we used a Bs4 promoter fragment of 302 bp (Schornack et al., 2005) in pENTR-D (Invitrogen). In the Bs4 UPAmut, all C nucleotides in the UPAAvrBs3 box are replaced by G nucleotides. The boxes were inserted via Phusion site-directed mutagenesis using primers Bs3inBs4-promfwd-PR (5′-CAATTTTATTATATAAACCTAACCATCCTCACAACGTTTCAAGTGGTACTTGT-3′) and Bs3ubm1inBs4-promfwd-PR (5′-CAATTTTATTATATAAAGGTAAGGATCCTCACAACGTTTCAAGTGGTACTTGT-3′) in combination with the primer Bs3in4-promrev-PR (5′-GTGAAAGCTTGTATTAACATTCGCTTTG-3′). After sequence analysis, the promoter constructs were transferred by Gateway LR recombination into the T-DNA vector pK7-GW-Bs3. pK7-GW-Bs3 was generated on the basis of pK7FWG2 (Karimi et al., 2002). We removed the HindIII and BamHI fragments by restriction digest from pK7FWG2 (contains the cauliflower mosaic virus 35S promoter, the Gateway attR cassette, and the GFP cds) and replaced it by a synthesized DNA fragment that contains SacI and EcoRV restriction sites followed by the cauliflower mosaic virus 35S terminator. Next, a Gateway attR cassette was placed into the EcoRV site, resulting in pK7-GW. The Bs3 cds was amplified from genomic DNA of pepper cv ECW-30R using the primers final-entry-SacI-01-fwd-PR (5′-GGGGGGAGCTCATGATGAATCAGAATTGCTTTAATTCTTGTTC-3′) and final-entry-SacI-02-rev-PR (5′-GGGGGGAGCTCCATTTGTTCTTTCCAAATTTTGGCAATATC-3′). These primers add a SacI restriction site on both ends of the Bs3 cds. The Bs3 cds was cloned into the SacI restriction site of pK7-GW, resulting in pK7-GW-Bs3.
Linker-Scanning Mutagenesis of the Bs3 Promoter
For insertion mutagenesis, we used the GPS-LS Linker Scanning System (New England Biolabs). As template, we used pENTR-D containing 343 bp 5′ of the ATG start codon of the Bs3 gene fused to the Bs3 cDNA. This plasmid was created by splicing using overlap extension PCR. The promoter was amplified from genomic DNA of ECW-30R pepper plants using primers Prom-356 bp-fwd-PR (5′-CACCTCATAGTCAAGCTAACGAAACTTATGC-3′) and B5-rev-PR (5′-CATACGGAACACTGTATTGCTTAAGG-3′). For cDNA synthesis, pepper ECW-30R plants were inoculated with a blunt syringe using Xanthomonas campestris pv vesicatoria strain 85-10 expressing avrBs3 (pDS300F; optical density at 600 nm [OD600] = 0.4). RNA extraction and cDNA synthesis were done as described previously (Römer et al., 2007). The Bs3 cDNA was amplified with Phusion high-fidelity DNA polymerase and the primers Final-entry-01-fwd-PR (5′-ATGATGAATCAGAATTGCTTTAATTCTTGTTC-3′) and Final-entry-01-rev-PR (5′-CTACATTTGTTCTTTCCAAATTTTGGCAATATCTTGTGC-3′). PCR products of the cDNA and the promoter region were mixed in a 1:1 ratio and PCR amplified using Prom-356 bp-fwd-PR and Final-entry-01-rev-PR primers. The PCR product was cloned into pENTR-D (Invitrogen), sequenced, and used for the transposon mutagenesis. Transposon insertions in the promoter region were identified via PCR. The identified transposon mutants were treated according to the manual, so that only the 15-bp insertion of the transposon was left. The resulting transposon mutants were sequenced and then recombined into pGWB1.
Plant Material and Infection Assays
Pepper and Nicotiana benthamiana plants were grown as described previously (Römer et al., 2007). Pepper germplasm was supplied by the U.S. Department of Agriculture (accessions preceded by “PI” or “Grif”) and the Plant Genetic Resources cluster of the Centre for Genetic Resources, The Netherlands (accessions preceded by CGN). Information on corresponding pepper accessions is available at http://www.ars-grin.gov/npgs/acc/acc_queries.html and http://www.cgn.wur.nl/UK/CGN+Plant+Genetic+Resources/. Agrobacterium-mediated transient transformation of N. benthamiana leaves, Xcv infection assays of Capsicum species, RT-PCR, RACE, and EMSA were carried out as described previously (Römer et al., 2007). Generally, Xanthomonas and Agrobacterium infection assays were routinely carried out at three independent time points. At each time point, each bacterial strain or combination of strains was inoculated into four different leaves. Agrobacterium strains delivering T-DNAs that encode bacterial effector genes were generally used in multiple infiltrations, always including an appropriate positive and negative control on each inoculated leaf.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide polymorphisms of Bs3 alleles.
Supplemental Figure S2. Proteins that are encoded by naturally occurring Bs3 alleles.
Supplemental Figure S3. Sequence comparison of TALe hypervariable residues.
Supplemental Figure S4. Comparison of UPAAvrBs3 boxes of different AvrBs3-inducible promoters.
Supplemental Figure S5. Alignment of UPAAvrBs3Δrep16 box variants in naturally occurring Bs3 alleles.
Supplemental Data Set S1. Bs3 promoter deletions.
Supplemental Data Set S2. Nucleotide sequences of linker-scanning Bs3 promoter mutants.
Supplemental Data Set S3. Nucleotide sequences of naturally occurring Bs3 alleles in Fasta format.
Supplemental Data Set S4. Sequence alignment of naturally occurring Bs3 alleles.
Supplementary Material
Acknowledgments
We are grateful to Diana Horvath, Annett Strauss, and Christoph Peterhänsel for helpful comments on earlier versions of the manuscript. We acknowledge the technical support of Sabine Recht, Jens Hausner, Carola Kretschmer, Bianca Rosinsky, and Marina Schulze. Seeds were obtained from the U.S. Department of Agriculture-Agricultural Research Service National Genetic Resources Program and the Centre for Genetic Resources (Wageningen University and Research Centre).
This work was supported by the Exzellenznetzwerk Biowissenschaften (Ministry of Culture of Saxonia-Anhalt), the 2Blades Foundation, and the Deutsche Forschungsgemeinschaft (grant nos. SFB 648, SPP 1212, and LA1338/2–2 to T.L. and grant no. SFB 648 to U.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Thomas Lahaye (lahaye@genetik.uni-halle.de).
The online version of this article contains Web-only data.
References
- Abramovitch RB, Anderson JC, Martin GB (2006) Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol 7 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballvora A, Pierre M, Van den Ackerveken G, Schornack S, Rossier O, Ganal M, Lahaye T, Bonas U (2001) Genetic mapping and functional analysis of the tomato Bs4 locus, governing recognition of the Xanthomonas campestris pv. vesicatoria AvrBs4 protein. Mol Plant Microbe Interact 14 629–638 [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45 399–436 [DOI] [PubMed] [Google Scholar]
- Block A, Li G, Fu ZQ, Alfano JR (2008) Phytopathogen type III effector weaponry and their plant targets. Curr Opin Plant Biol 11 396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas U, Stall RE, Staskawicz B (1989) Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol Gen Genet 218 127–136 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Chandler JW (2009) Local auxin production: a small contribution to a big field. Bioessays 31 60–70 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814 [DOI] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, et al (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20 1250–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S, Lindeberg M, Collmer A (2009) Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr Opin Microbiol 12 53–60 [DOI] [PubMed] [Google Scholar]
- Göhre V, Robatzek S (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol 46 189–215 [DOI] [PubMed] [Google Scholar]
- Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL (2006) Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol 60 425–449 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Yao N (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6 201–211 [DOI] [PubMed] [Google Scholar]
- Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, et al (2005) R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435 1122–1125 [DOI] [PubMed] [Google Scholar]
- Herbers K, Conrads-Strauch J, Bonas U (1992) Race-specificity of plant resistance to bacterial spot disease determined by repetitive motifs in a bacterial avirulence protein. Nature 356 172–174 [Google Scholar]
- Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S (2009) Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22 115–122 [DOI] [PubMed] [Google Scholar]
- Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inzé D, Engler G, Villarroel R, et al (1980) The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid 3 212–230 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Kamoun S (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol 44 41–60 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Kay S, Bonas U (2009) How Xanthomonas type III effectors manipulate the host plant. Curr Opin Microbiol 12 37–43 [DOI] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marois E, Hause G, Bonas U (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318 648–651 [DOI] [PubMed] [Google Scholar]
- Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, Lee SY, Blakeslee JJ, Murphy AS, Bohnert HJ, et al (2007) yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol 145 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5 305–315 [DOI] [PubMed] [Google Scholar]
- McKnight SL, Kingsbury R (1982) Transcriptional control signals of a eukaryotic protein-coding gene. Science 217 316–324 [DOI] [PubMed] [Google Scholar]
- Minsavage GV, Dahlbeck D, Whalen MC, Kearny B, Bonas U, Staskawicz BJ, Stall RE (1990) Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria-pepper interactions. Mol Plant Microbe Interact 3 41–47 [Google Scholar]
- Mudgett MB (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol 56 509–531 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104 34–41 [DOI] [PubMed] [Google Scholar]
- Römer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T (2007) Plant-pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318 645–648 [DOI] [PubMed] [Google Scholar]
- Schlaich NL (2007) Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci 12 412–418 [DOI] [PubMed] [Google Scholar]
- Schornack S, Ballvora A, Gürlebeck D, Peart J, Baulcombe D, Baker B, Ganal M, Bonas U, Lahaye T (2004) The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J 37 46–60 [DOI] [PubMed] [Google Scholar]
- Schornack S, Meyer A, Römer P, Jordan T, Lahaye T (2006) Gene-for-gene mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J Plant Physiol 163 256–272 [DOI] [PubMed] [Google Scholar]
- Schornack S, Minsavage GV, Stall RE, Jones JB, Lahaye T (2008) Characterization of AvrHah1 a novel AvrBs3-like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol 179 546–556 [DOI] [PubMed] [Google Scholar]
- Schornack S, Peter K, Bonas U, Lahaye T (2005) Expression levels of avrBs3-like genes affect recognition specificity in tomato Bs4 but not in pepper Bs3 mediated perception. Mol Plant Microbe Interact 18 1215–1225 [DOI] [PubMed] [Google Scholar]
- Sugio A, Yang B, Zhu T, White FF (2007) Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proc Natl Acad Sci USA 104 10720–10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurek B, Marois E, Bonas U, Van den Ackerveken G (2001) Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J 26 523–534 [DOI] [PubMed] [Google Scholar]
- Van den Ackerveken G, Marois E, Bonas U (1996) Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87 1307–1316 [DOI] [PubMed] [Google Scholar]
- Wichmann G, Bergelson J (2004) Effector genes of Xanthomonas axonopodis pv. vesicatoria promote transmission and enhance other fitness traits in the field. Genetics 166 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF (2006) Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci USA 103 10503–10508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zhu W, Johnson LB, White FF (2000) The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent nuclear-localized double-stranded DNA-binding protein. Proc Natl Acad Sci USA 97 9807–9812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.