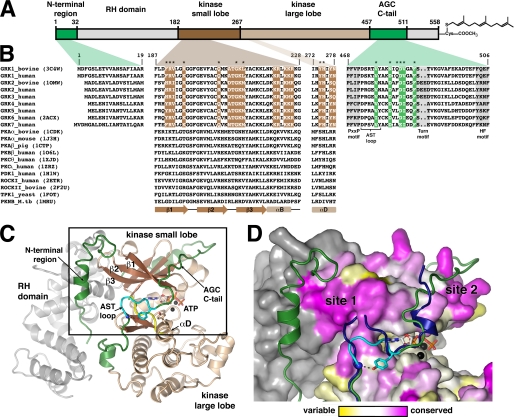
FIGURE 1.
GRK surface residues potentially important for GPCR phosphorylation. A, domain architecture of bGRK1. B, sequence alignment of regions from GRKs that were targeted in this study with other AGC kinases. Colored boxes map these regions back to the domain structure shown in A. Regions of the core kinase domain that contain residues conserved in the GRK subfamily, but not in the extended AGC kinase family, are highlighted in brown. Conserved residues of the AGC kinase C-tail are highlighted in green. Positions investigated in this study are indicated with asterisks. Only one PDB accession code for each kinase of known structure is shown in parentheses. Residue numbers correspond to those of bGRK1. The PXXP, turn, and hydrophobic (HF) motifs (highlighted in gray) are characteristic features found in most AGC kinase C-tails (22). Yeast, Saccharomyces cerevisiae; M.tb, Mycobacterium tuberculosis. C, ribbon diagram of the bGRK1535-H6·ATP complex. The model is a hybrid that contains all the ordered elements from the two unique chains resolved in the crystal structure (PDB accession number 3C4W), such that the observed N terminus and a nearly complete AST region of the kinase C-tail are displayed in a single model. The RH domain is colored gray, and the β-strands and α-helices of the core kinase domain are dark and light brown, respectively. The hinge region joining the kinase small and large lobes (between β5 and αD) is colored yellow. The N-terminal region and the AGC kinase C-tail are shown in green and the AST loop in cyan. The ATP molecule bound in the active site is shown as a stick model, and the two associated Mg2+ ions are colored black. D, conservation scores of GRKs mapped onto the surface of bGRK1. The area boxed in C is shown. The conservation scores were calculated by comparing the sequence conservation of residues from the core kinase domain of GRKs with those of the entire AGC family (see text). Residues are colored using a gradient, from magenta (more conserved in GRKs than the AGC kinases) to white (as conserved in GRKs as in AGC kinases) and to yellow (more variable in GRKs than in AGC kinases). The RH domain, which was not included in this calculation, is colored gray. Highest conservation among GRKs cluster at two sites, “site 1” and “site 2.” Site 1 corresponds to a region of the small lobe left exposed by the shorter AST loop found in GRKs relative to other AGC kinases. The AST region of protein kinase B (PDB accession number 1O6K) is superimposed for comparison (blue ribbon).