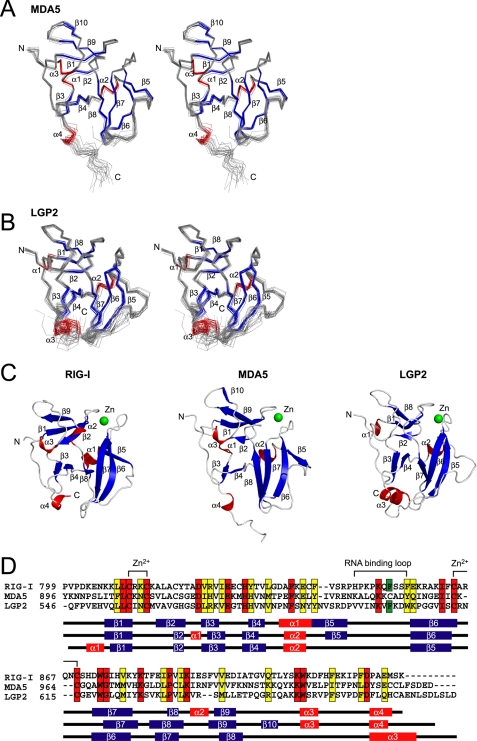
FIGURE 2.
Solution structure of MDA5 CTD and LGP2 CTD. A and B, best fit superposition of the backbone atoms of 20 NMR-derived MDA5 CTD (A) and LGP2 CTD (B). Structures are shown in stereo. β-Strands and α-helices are shown in blue and red, respectively. C, ribbon diagrams of the structure of RIG-I CTD, MDA5 CTD, and LGP2 CTD (left to right). Secondary structure elements are labeled. The figure was prepared using PyMOL. D, sequence alignment of human RIG-I, MDA5, and LGP2 CTDs. ClustalX was used to align the sequences. The secondary structure elements of each CTD are indicated below the alignment. The amino acids in red and yellow indicate conserved (red) and type-conserved (yellow) residues with the Zn2+ binding Cys-X-X-Cys motifs and RNA binding loop. The Phe residues conserved in RIG-I and LGP2 in the RNA binding loop are colored green.