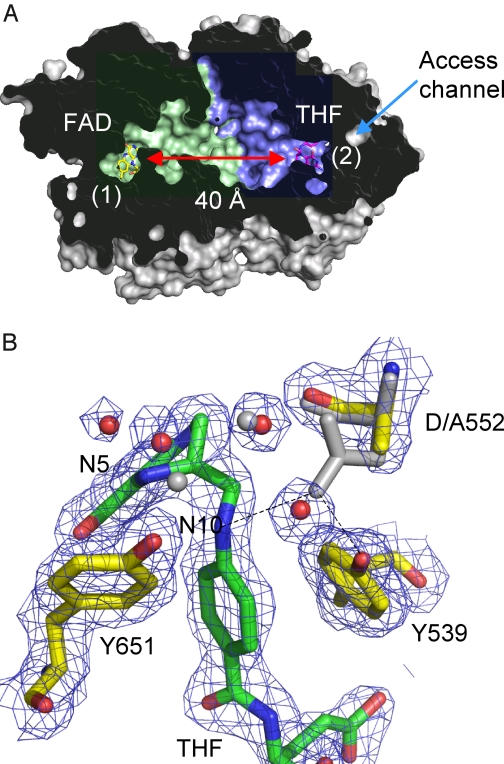
FIGURE 2.
DMGO structure. A, cross-sectional view of a DMGO monomer solvent-accessible surface, with the internal cavity colored blue for the FAD domain and green for the THF-binding domain (Ref. 22; Protein Data Bank code 3GSI). FAD and THF are shown as sticks designating the respective active sites (labeled 1 and 2), which are separated by ∼40 Å. The only access channel to the internal cavity coincides with the THF-binding site. B, detailed view of the THF-binding site for the D552A mutant crystal structure (Protein Data Bank code 3GSI). 2 Fo− Fc sigmaA-weighted electron density is shown as a blue mesh contoured at 1 σ surrounding THF and key amino acids. These are shown in atom-colored sticks (THF, green carbons; amino acids, yellow carbons) in addition to selected water molecules shown as red spheres. The corresponding wt DMGO structure is shown as gray sticks and spheres. This figure was made using PyMOL (DeLano Scientific).