FIGURE 4.
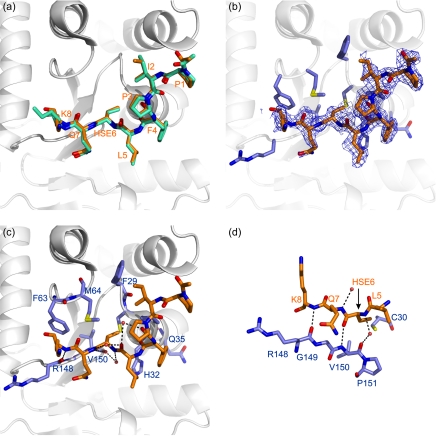
The substrate binding site of DsbA. a, comparison of the peptide conformation in the two models of the EcDsbA-peptide complex (Protein Data Bank code 3DKS). Peptide chains E and F are shown in orange and green stick representation, respectively, and the residues are labeled in orange. The EcDsbA molecule to which peptide E is covalently attached (chain C) is shown in a schematic diagram. b, omit (2Fo − Fc) electron density map (contoured at 0.8 σ) for the peptide (chain E). EcDsbA residues in contact with the peptide are shown in a blue stick representation. c, polar interactions observed in the complex are shown as black dotted lines between the interacting residues. Water atoms are shown as red spheres. The EcDsbA-peptide complex (chains C and E) is shown in the same orientation as above, and EcDsbA residues are labeled in blue. d, the peptide forms an antiparallel interaction with residues in the cis-proline loop of EcDsbA. Polar interactions are shown as black dotted lines between the peptide (in orange) and EcDsbA (in blue).