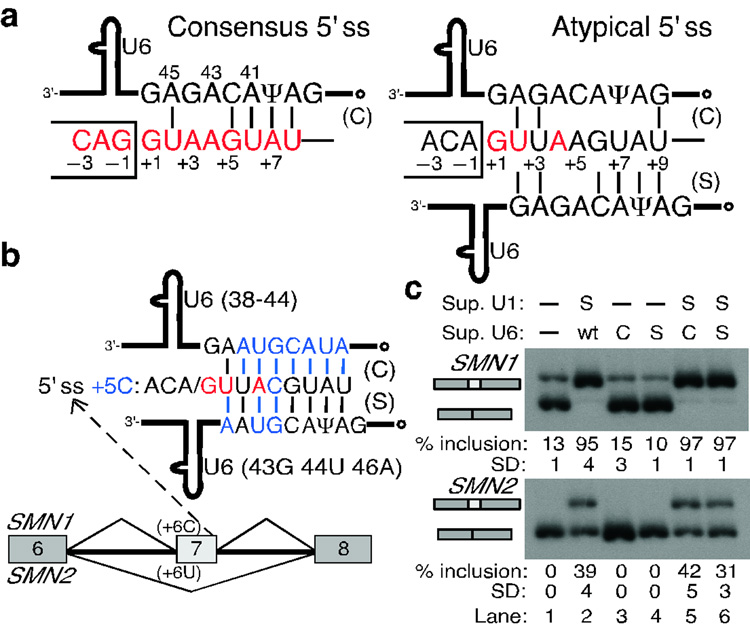
Figure 5. U6 snRNA does not base-pair to the atypical 5' ss in a shifted register.
a, Schematic of the base-pairing between consensus (left) or atypical (right) 5' ss and the conserved U6 ACAGAG box (positions are numbered). The open dot indicates the γ-monomethyl cap. The atypical 5' ss has an extended base-pairing potential to U6 in the shifted register. b, Schematic of the suppressor U6 snRNAs carrying compensatory mutations in either the canonical (C) or the shifted (S) register. These mutations (blue font) restore base-pairing for the +5C mutation at atypical 5' ss in the SMN1/2 context. c, RT-PCR analysis of the SMN1/2 minigenes cotransfected with suppressor U1 and U6 snRNAs. Top labels indicate the suppressor U1/U6 used. wt, wild-type U6 snRNA. Suppressor U6s alone had no effect (lanes 3, 4 vs. lane 1). In combination with suppressor U1, suppressor U6 in the canonical register resulted in more exon 7 inclusion than suppressor U6 in the shifted register (lanes 5 and 6 in SMN2). These results suggest that atypical 5' ss establish canonical base-pairing to U6 snRNA.