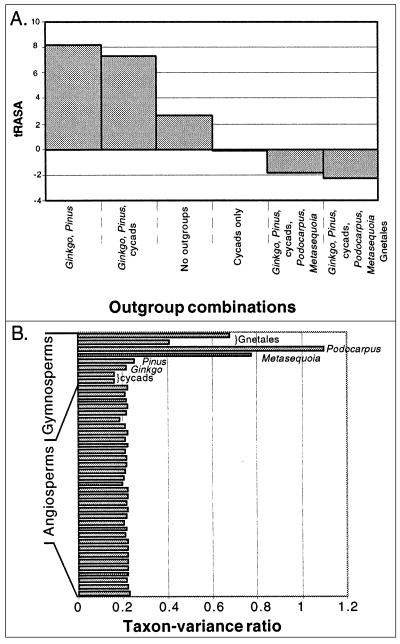
Figure 2.
(A) Optimal outgroup analyses (43) using the six-gene dataset indicate that Ginkgo and Pinus are the best species to use to identify the root node of angiosperm phylogeny. (B) Taxon-variance ratio plot showing that Gnetales (Gnetum and Welwitschia), Podocarpus, and Metasequoia are outliers relative to all other sampled taxa. Taxon-variance outliers have been shown to lead to inconsistent tree estimation when included in phylogenetic analyses (44).