Abstract
To gain a better understanding of the gene expression changes that occurs during sepsis, we have performed a cDNA microarray study utilizing a tissue culture model that mimics human sepsis. This study utilized an in vitro model of cultured human fetal cardiac myocytes treated with 10% sera from septic patients or 10% sera from healthy volunteers. A 1700 cDNA expression microarray was used to compare the transcription profile from human cardiac myocytes treated with septic sera vs normal sera. Septic sera treatment of myocytes resulted in the down-regulation of 178 genes and the up-regulation of 4 genes. Our data indicate that septic sera induced cell cycle, metabolic, transcription factor and apoptotic gene expression changes in human myocytes. Identification and characterization of gene expression changes that occur during sepsis may lead to the development of novel therapeutics and diagnostics.
Keywords: Septic sera, gene expression modulation, human myocytes
Introduction
Septic shock (shock due to infection) and sepsis associated multiple organ failure are the number one cause of death in North American intensive care units with an incidence which continues to increase [1, 2]. Approximately 800,000 cases of sepsis are admitted every year to hospitals in North America and despite aggressive antibiotics and supportive care, over 200,000 patients per year succumb to this disorder [1]. The typical human cardiovascular response to septic shock is characterized by hypotension, decreased systemic vascular resistance and elevated cardiac index. In addition, myocardial depression manifested by reversible biventricular dilation and reduction of ejection fraction has been shown to be common in spontaneous human septic shock [3, 4]. Deaths are typically due to early refractory cardiovascular failure (hypotension or shock) or later multiple organ failure. Shock and organ failure may occur and progress to death despite the fact that complete eradication of the invading organism can usually be achieved with antibiotic support [3, 4]. Severe infection is able to initiate a conserved inflammatory genetic/metabolic cascade in the host that progresses to organ injury, organ system dysfunction and death despite elimination of the initial trigger.
Elevated levels of various cytokines have been observed in sera from patients with sepsis and septic shock [5]. Tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β) [6] and migration inhibitory factor (MIF) [7], play a critical role in the development of cellular dysfunction observed during sepsis [8]. Multiple processes have been identified in mediating myocardial dysfunction which include; apoptosis [9-11], mitochondrial dysfunction [12, 13], alterations in calcium current, decrease in myofilament calcium sensitivity [14, 15], autonomic dysfunction due to neuronal apoptosis [16], metabolic alterations [17-19] and inducible nitric oxide synthase (iNOS) induction [20].
The high-throughput of DNA microarrays allows for the monitoring of thousands of genes and the identification of transcriptional changes that occur in gene expression. In a novel approach to diagnose sepsis, a recent study described a distinct expression profile of patients with early sepsis that differs from critically ill systemic inflammatory response syndrome (SIRS) patients [21]. Tang et al [22] provided data which was used to distinguish between sepsis and SIRS patients by utilizing neutrophil gene expression profiling.
In this study, we used a 1700 human cDNA microarray to analyze the gene expression patterns in a tissue culture model which mimics human sepsis. In this model human fetal cardiac myocytes were incubated for 12 hours with 10% human septic sera and 10% sera from healthy volunteers. We showed that human fetal cardiac myocytes responded to human septic serum through the repression of 178 genes and the up-regulation of 4 genes. Septic sera treatment of human myocytes induced the differential expression of several metabolic, transcriptional, cell cycle and developmental genes that contribute to cellular dysfunction observed in septic patients.
Materials and methods
Study patients from whom human septic sera was obtained
Serum was derived from 4 patients who were in acute phase of septic shock as defined by modified ACCP/SCCM Consensus Conference criteria [23]. Patients were required to have all (rather than a minimum of two) of the following four criteria for systemic inflammatory response syndrome 1) a body temperature greater than 38°C or less than 36°C; 2) a heart rate greater than 90 beats per minute; 3) tachypnea, manifested by a respiratory rate greater than 20 breaths per minute or hyperventilation, as indicated by a PaCO2 of less than 32 mm Hg; 4) an alteration in the white blood cell count (i.e. WBC greater than 12,000/ mm3, less than 4,000/mm3, or the presence of more than 10% immature neutrophils). In addition, all patients whose serum samples were utilized exhibited positive blood cultures with a defined focus of infection (e.g. peritonitis or pneumonia) and required substantial pressor therapy (>0.5 ug/kg/min norepinephrine) to maintain mean arterial pressure > 65 mm Hg. Serum samples were obtained within 24 hours of presentation with septic shock. Human septic serum was obtained after informed consent under an approved Institutional Review Board (Rush University) approved protocol. Subjects contributing human septic sera for this study were not known to have pre-existing structural heart disease. 10 cc of blood was drawn from the patient and centrifuged for 10 min at 1700×g. The supernatant representing the serum was aliquoted and stored at −70°C. Key characteristics and cytokine profiles of sera donors with septic shock is shown in Table 1. Normal human sera was harvested from healthy lab volunteers.
Table 1.
Subject information, infecting organism and serum cytokine concentrations (TNF-α, IL-1β, IFN-γ)
| Patient | Gender | Age | Infecting organism | Outcome | TNF-α (pg/mL) | IL-1β (pg/mL) | IFN-γ (pg/mL) |
|---|---|---|---|---|---|---|---|
| SE | Female | 81 | Staphylococcus aureus | Died | 43.9 | 6.1 | 22.9 |
| SF | Male | 73 | Heamophilus influenzae | Survived | 13.4 | 3.6 | 6.8 |
| SG | Male | 68 | Escherichia coli | Survived | 19.0 | 7.7 | 10.3 |
| SH | Male | 54 | Streptococcus pneumoniae | Survived | 39.0 | 1.8 | 6.0 |
| Normal | Healthy individuals | — | — | — |
Data are for the 4 septic individuals and the healthy volunteers. Cytokine concentrations were determined using a sandwich ELISA
Sandwich ELISA assays
Human septic and normal sera concentrations of TNF-α, IL-1β and IFN-γ were determined by ELISA. For the detailed protocol, refer to the DuoSet ELISA Development System on the website www.RnDSystems.com or in the following product manuals: Human TNF-α/TNFSF1A, Catalog Number: DY210; Human IL-1β/IL-1F2, Catalog Number: DY201; and Human IFN-γ, Catalog Number: DY285. The following modifications were made to the ELISA protocol. Four washes were performed instead of three in order to reduce the background. Furthermore, the reagent diluents were optimized for the quantification of each cytokine. The diluents consisted of 10% FCS with PBS, 2% FCS with PBS and 0.5% FCS with PBS for TNF-α, IL-1β and IFN-γ, respectively.
Treatment of human myocytes with human septic sera
Human fetal myocytes (ScienCell Research) were grown to 2 × 106 cells per 10 cm plate and treated with either 10% septic sera or 10% normal sera.
Total RNA extraction and cDNA synthesis
Total RNA was extracted from all cultured human fetal cardiac myocyte cells using a Qiagen RNA miniprep kit (Qiagen, Hilden, Germany). Isolated RNA was used as a template for cDNA generation using reverse transcriptase. RNA was gel checked for integrity.
Microarray preparation
cDNA preparations from both normal and septic sera treated cardiac myocytes were labeled with either Cy3- or Cy5-conjugated deoxyribonucleoside triphosphates, and were arbitrary matched and allowed to hybridize to microarray slides as described previously [24]. Changes in gene expression in fetal cardiac myocytes responding to septic serum were detected using 1.7k human cDNA microarray (University Health Network Microarray Center, Toronto, ON, Canada).
Data analysis
Hybridized and washed arrays were scanned on an Axon 4000B dual laser scanner (532 nm/633 nm wavelengths) as described previously [25]. The fluorescence intensities for each feature (spot) on the array were determined using GenePix Pro array software (version 3.0) from Axon Instruments. The GenePix result files (.gpr) were imported into Microsoft Excel worksheets (.xls). The data from duplicate spots on the 8 independent array experiments (4 with reverse fluorescence) were corrected for background and normalized using a previously described method [26]. Filtering of significant genes was performed using a confidence matrix that compared the average ratio between septic and normal sera for each individual from both groups and the total ratio average between the septic group and the healthy individual group. A heat map of this gene list was created using the TM4 microarray software suite version 2.19 [27]. Functional gene classification and ontology analyses were performed by uploading specific gene expression lists to the web-based application D.A.V.I.D. (Database for Annotation, Visualization and Integrated Discovery) [28].
Real-time PCR
Verification of the differential gene expression results detected by the cDNA microarray was conducted using Q-PCR. Duplicate Q-PCR reactions were performed as described previously [25]. A 6 point standard curve was constructed for each gene. GAPDH was used as the reference gene. The gene-specific oligo-nucleotide primers to confirm the array data using Q-PCR were: API5: forward primer, 5′-CGACAGTAGAGGAGCTTTACCG-3′, reverse primer, 5′-GCTGCTAATCGCTTTTCCTTAGT-3′; FECH: forward primer, 5′-GTGGAGCACTATTGACAGGTG-3′, reverse primer, 5′-CCACAGACATCGGCAGTGA-3′; STMN1: forward primer, 5′-GCCCTCGGTCAAAAGAATCTG-3′, reverse primer, 5′-TGCTTCAAGACCTCAGCTTCA-3′; GADD45A: forward primer, 5′-GAGAGCAGAAGACCGAAAGGA-3′, reverse primer, 5′-CACAACACCACGTTATCGGG-3′; RGS4: forward primer, 5′-CAAGCCGGAACATGCTAGAG-3′, reverse primer, 5′CGGGTTGACCAAATCAAGATAGA-3′; GAPDH: forward primer, 5′-CATGAGAAGTATGACAACAGCCT-3′, reverse primer, 5′-AGTCCTTCCACGATACCAAAGT-3′. To verify the changes in gene expression detected in fetal cardiac myocytes treated with serum from septic patients, we conducted Q-PCR analysis on five differentially expressed transcripts. RNA was isolated from fetal cardiac myocytes treated with either 10% serum from healthy volunteers or 10% human septic serum, reverse transcribed, cDNA was used in the Q-PCR reaction using the previously described reaction [25]. Using gene-specific primers for API5, STMN1, FECH, RGS4 and GADD45A, the relative expression of these genes to the housekeeping gene GAPDH was calculated. Fold change in gene expression in septic sera treated samples as compared to normal sera treated samples was consistent with the changes as detected by the microarray analysis. Q-PCR showed an increase in the expression of API5, STMN1, and FECH and a decrease in the expression of RGS4 and GADD45A.
Results and discussion
Sera Cytokine Analysis
The levels of TNF-α, IL-1β and IFN-γ in the sera of the four septic patients and in the serum of healthy individuals were quantified using the Enzyme-Linked Immunosorbant Assay (ELISA) kit from R&D Systems. Patient SE was a female of 81 years old that had Staphylococcus aureus as the infecting organism during sepsis. Patient SE succumbed to the disease. The levels of TNF-α, IL-1β and IFN-γ for this patient were 43.9 pg/ml, 6.1 pg/ml and 22.8 pg/ml respectively (Table 1). Patient SF a male of 73 years, with the infecting organism Haemophilus influenzae survived from sepsis. This patient had levels of TNF-α, IL-1β and IFN-γ of 13.4 pg/ml, 3.6 pg/ml and 6.8 pg/ml respectively. Patient SG was infected with Escherichia coli. This patient, a male of 68 years old also survived sepsis and the levels of cytokines measured from the serum of this patient were 19.0 pg/ml, 7.7 pg/ml and 10.3 pg/ml for TNF-α, IL-1β and IFN-γ respectively. The fourth serum SH was from a male of 54 years of age with Staphylococcus pneumonia as the initial infecting organism for sepsis. This patient also survived sepsis and had levels of 39.0 pg/ml, 1.8 pg/ml and 6.0 pg/ml for TNF-α, IL-1β and IFN-γ in his serum. Normal sera from healthy lab volunteers did not have measurable levels of TNF-α, IL-1β and IFN-γ.
Modulation of gene expression
We have shown that the human fetal cardiac myocytes responded to human septic sera through the repression of 178 genes and the up-regulation of 4 genes (Table 2 and see the corresponding heat map Figure 1). The observed general down regulation of gene expression in response to septic sera may be attributed to the elevated levels of the cytokines, TNF-α, IFN-γ and IL-1β (Table 1) in the septic sera which may be inducing cellular dysfunction in the myocytes. Elevated levels of cytokines in septic sera have been previously shown to induce cellular dysfunction in human myocytes [29]. A key element of this tissue culture model of sepsis is that septic myocardial depression induced by septic serum has been shown to correlate with the amount of myocardial depression seen in intact humans using radionuclide measures of ejection fraction [30].
Table 2.
Fold change in gene expression of septic to normal sera treated cardiac myocytes using the 1.7K human cDNA array
| Genbank Accession ID | Gene Symbol | Description | Fold Change |
|---|---|---|---|
| Apoptosis | |||
| AA136799 | GADD45A | growth arrest and DNA-damage-inducible, alpha | 0.28 |
| N34233 | MAGEH1 | melanoma antigen family H, 1 | 0.33 |
| BG620906 | CSE1L | CSE1 chromosome segregation 1-like (yeast) | 0.46 |
| T78285 | CRADD | CASP2 and RIPK1 domain containing adaptor with death domain | 0.51 |
| N91060 | VEGF | vascular endothelial growth factor | 0.53 |
| AA011445 | CASP3 | caspase 3, apoptosis-related cysteine peptidase | 0.59 |
| W92108 | DHCR24 | 24-dehydrocholesterol reductase | 0.60 |
| H79188 | ERCC2 | excision repair cross-complementing rodent repair deficiency, complementation group 2 (xeroderma pigmentosum D) | 0.65 |
| BI824641 | BTK | Bruton agammaglobulinemia tyrosine kinase | 0.70 |
| BG387747 | API5 | apoptosis inhibitor 5 | 2.21 |
| Catalysts | |||
| W31103 | BRD2 | bromodomain containing 2 | 0.53 |
| AA039228 | FNTB | farnesyltransferase, CAAX box, beta | 0.54 |
| H11807 | CTPS | CTPsynthase | 0.57 |
| H62727 | CRYM | crystallin, mu | 0.58 |
| N77157 | PTPN12 | protein tyrosine phosphatase, non-receptor type 12 | 0.61 |
| AA031513 | MMP7 | matrix metallopeptidase 7 (matrilysin, uterine) | 0.64 |
| BE738657 | PBEF | pre-B-cell colony enhancing factor 1 | 0.65 |
| N50000 | MAT1A | methionine adenosyltransferase I, alpha | 0.65 |
| W92066 | RFC4 | replication factor C (activator 1) 4, 37kDa | 0.66 |
| Cell adhesion | |||
| H29191 | APC | adenomatosis polyposis coli | 0.08 |
| BI496175 | ALCAM | activated leukocyte cell adhesion molecule | 0.47 |
| H16046 | ITGA6 | integrin, alpha 6 | 0.52 |
| H30141 | SELP | selectin P (granule membrane protein 140kDa, antigen CD62) | 0.60 |
| T80274 | CD34 | CD34 antigen | 0.64 |
| W01300 | ITGB3 | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | 0.66 |
| Cell cycle | |||
| T65624 | DUSP6 | dual specificity phosphatase 6 | 0.16 |
| H71112 | MCM2 | MCM2 minichromosome maintenance deficient 2, mitotin (S. cerevisiae) | 0.46 |
| H14471 | MCM6 | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 0.46 |
| BG424365 | SES2 | sestrin 2 (SESN2) | 0.52 |
| H04421 | DUSP1 | dual specificity phosphatase 1 | 0.55 |
| AA001329 | CCNA2 | cyclin A2 | 0.62 |
| BG403841 | NHP2L1 | NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) | 0.62 |
| R14363 | HDAC5 | histone deacetylase 5 | 1.88 |
| Cell proliferation | |||
| AA128253 | PMP22 | peripheral myelin protein 22 | 0.30 |
| AI819719 | PDGFRA | platelet-derived growth factor receptor, alpha polypeptide | 0.30 |
| AA114957 | MXI1 | MAX interactor 1 | 0.45 |
| W95000 | CDC25C | cell division cycle 25C | 0.47 |
| W47595 | TGFB2 | transforming growth factor, beta 2 | 0.48 |
| AA028183 | TGFBR2 | transforming growth factor, beta receptor II (70/80kDa) | 0.53 |
| H29950 | TOB1 | transducer of ERBB2, 1 | 0.58 |
| BG565696 | PRDX1 | peroxiredoxin 1 | 0.60 |
| AA055757 | CSRP2 | cysteine and glycine-rich protein 2 | 0.60 |
| AA037107 | TGFA | transforming growth factor, alpha | 0.60 |
| AA044049 | GNRH1 | gonadotropin-releasing hormone 1 | 0.61 |
| H12419 | USP8 | ubiquitin specific peptidase 8 | 0.63 |
| W17355 | DTR | heparin-binding EGF-like growth factor (HBEGF) | 0.63 |
| AA007492 | RBBP4 | retinoblastoma binding protein 4 | 0.64 |
| W19744 | CDK6 | Cyclin-dependent kinase 6 (CDK6) | 0.67 |
| W78793 | EPS8 | epidermal growth factor receptor pathway substrate 8 | 0.69 |
| Cellular component organization and intracellular transport | |||
| AA056151 | SEC24C | SEC24 related gene family, member C (S. cerevisiae) | 0.27 |
| BG753663 | TUBB5 | tubulin, beta 5 | 0.52 |
| H59171 | SEC23B | Sec23 homolog B (S. cerevisiae) | 0.52 |
| W01720 | SRPR | signal recognition particle receptor (‘docking protein’) | 0.57 |
| W92260 | RAB11A | RAB11A, member RAS oncogene family | 0.58 |
| W33064 | TUBA1 | tubulin, alpha 1 (testis specific) | 0.58 |
| BG169044 | ARL6IP | ADP-ribosylation factor-like 6 interacting protein | 0.58 |
| W48577 | GARS | glycyl-tRNAsynthetase | 0.60 |
| R20063 | TPR | translocated promoter region (to activated MET oncogene) | 0.62 |
| W16514 | RHO6 | Rho family GTPase 1 (RND1) | 0.63 |
| AA053988 | ARF1 | ADP-ribosylation factor 1 | 0.64 |
| N31521 | BPAG1 | dystonin (DST) | 0.64 |
| BG770889 | Rab11-FIP2 | RAB11 family interacting protein 2 (class I) | 0.64 |
| H28534 | AQP1 | aquaporin 1 (channel-forming integral protein, 28kDa) | 0.64 |
| N34169 | NF2 | neurofibromin 2 (bilateral acoustic neuroma) | 0.65 |
| Developmental processes | |||
| BG740719 | DDX5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 0.31 |
| AA037738 | HGF | hepatocyte growth factor (hepapoietin A; scatter factor) | 0.38 |
| H09372 | PPP2CA | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | 0.41 |
| T75267 | FRAP1 | FK506 binding protein 12-rapamycin associated protein 1 | 0.43 |
| N40420 | CCND1 | cyclin D1 | 0.51 |
| W48569 | BDNF | brain-derived neurotrophic factor | 0.52 |
| H52752 | HSPA9B | heat shock 70kDa protein 9B (mortalin-2) | 0.55 |
| W49766 | WNT5A | wingless-type MMTV integration site family, member 5A | 0.55 |
| W01469 | ETS2 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | 0.61 |
| N25434 | MSX2 | msh homeo box homolog 2 (Drosophila) | 0.62 |
| W38673 | ID2 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 0.63 |
| BI198063 | IGFBP2 | insulin-like growth factor binding protein 2, 36kDa | 0.67 |
| DNA Repair | |||
| AA134026 | UBE2A | ubiquitin-conjugating enzyme E2A (RAD6 homolog) | 0.41 |
| T78280 | HAT1 | histone acetyltransferase 1 | 0.43 |
| AA028978 | ERCC5 | excision repair cross-complementing rodent repair deficiency, complementation group 5 (xeroderma pigmentosum, complementation group G (Cockayne syndrome)) | 0.56 |
| H30857 | MLH1 | mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) | 0.63 |
| Integral to plasma memebrane | |||
| H29706 | GPC5 | glypican 5 | 0.37 |
| W05301 | CD1E | CD1E antigen, e polypeptide | 0.55 |
| BF212425 | RAI3 | G protein-coupled receptor, family C, group 5, member A (GPRC5A) | 0.59 |
| Metabolism | |||
| BG762226 | PCSK2 | proprotein convertase subtilisin/kexin type 2 | 0.40 |
| N63511 | DPYD | dihydropyrimidine dehydrogenase | 0.55 |
| T99689 | MTR | 5-methyltetrahydrofolate-homocysteine methyltransferase | 0.55 |
| H15303 | PCCB | propionyl Coenzyme A carboxylase, beta polypeptide | 0.57 |
| H19387 | ME2 | malic enzyme 2, NAD(+)-dependent, mitochondrial | 0.57 |
| BI193156 | SDF2 | stromal cell-derived factor 2 | 0.58 |
| AA029397 | PSMA6 | proteasome (prosome, macropain) subunit, alpha type, 6 | 0.58 |
| BE254368 | FDPS | farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) | 0.59 |
| AA029803 | HDLBP | high density lipoprotein binding protein (vigilin) | 0.59 |
| BI160119 | RPL27A | ribosomal protein L27a | 0.59 |
| R98094 | BPGM | 2,3-bisphosphoglycerate mutase | 0.60 |
| R52371 | SLC35A1 | solute carrier family 35 (CMP-sialic acid transporter), member A1 | 0.61 |
| H30710 | BAT1 | HLA-B associated transcript 1 | 0.63 |
| R01732 | AMPD3 | adenosine monophosphate deaminase (isoform E) | 0.63 |
| W68191 | CLU | clusterin (complement lysis inhibitor, SP-40,40, sulfated glycoprotein 2, testosterone-repressed prostate message 2, apolipoprotein J) | 0.63 |
| N31054 | MPV17 | MpV17 transgene, murine homolog, glomerulosclerosis | 0.64 |
| W94208 | FDFT1 | farnesyl-diphosphate farnesyltransferase 1 | 0.64 |
| W92014 | ADH6 | alcohol dehydrogenase 6 (class V) | 0.64 |
| W52843 | ALDH3B1 | aldehyde dehydrogenase 3 family, member B1 | 0.65 |
| W31637 | LPL | lipoprotein lipase | 0.65 |
| H82585 | NT5E | 5′-nucleotidase, ecto (CD73) | 0.65 |
| W37213 | HEM1 | NCK-associated protein 1-like (NCKAP1L) | 0.66 |
| Metal binding | |||
| H93255 | MT1E | metallothionein 1E (functional) | 0.62 |
| R18562 | ATP7B | ATPase, Cu++ transporting, beta polypeptide | 0.62 |
| R99208 | MT1B | metallothionein 1B (functional) | 0.64 |
| Mitochondrial | |||
| W37672 | HK1 | hexokinase 1 | 0.60 |
| AA133258 | HADHB | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 0.62 |
| W21228 | UQCRC2 | ubiquinol-cytochrome c reductase core protein II | 0.65 |
| N72504 | FECH | ferrochelatase (protoporphyria) | 2.4 |
| Nervous System(Development) | |||
| R15219 | EPHA7 | EPH receptor A7 (EPHA7) | 0.45 |
| H17696 | MBP | myelin basic protein | 0.46 |
| BI826246 | PLP1 | proteolipid protein 1 (Pelizaeus-Merzbacher disease, spastic paraplegia 2, uncomplicated) | 0.52 |
| Protein (folding/unfolding/proteolysis) | |||
| H18866 | CHRM3 | cholinergic receptor, muscarinic 3 | 0.31 |
| H03673 | KRT7 | keratin 7 | 0.48 |
| AA036988 | CCT3 | chaperonin containing TCP1, subunit 3 (gamma) | 0.53 |
| H62639 | HSPA8 | heat shock 70kDa protein 8 | 0.56 |
| W90284 | HERPUD1 | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | 0.57 |
| H53489 | C4BPA | complement component 4 binding protein, alpha | 0.62 |
| R48054 | CCT5 | chaperonin containing TCP1, subunit 5 (epsilon) | 0.62 |
| AA031513 | MMP7 | matrix metallopeptidase 7 (matrilysin, uterine) | 0.64 |
| T74000 | BLMH | bleomycin hydrolase | 0.64 |
| Regulation of Translation (Initiators/Inhibitors) | |||
| H79856 | UK114 | (HRSP12) heat-responsive protein 12 | 0.29 |
| AA098888 | ETF1 | eukaryotic translation termination factor 1 | 0.58 |
| N79725 | MTIF2 | mitochondrial translational initiation factor 2 | 0.58 |
| BG178139 | WBSCR1 | Williams-Beuren syndrome chromosome region 1 | 0.63 |
| Signaling | |||
| T74308 | MAPK6 | mitogen-activated protein kinase 6 | 0.20 |
| AI267373 | RGS4 | Regulator of G-protein signalling 4 (RGS4) | 0.28 |
| H18866 | CHRM3 | cholinergic receptor, muscarinic 3 | 0.31 |
| AA055349 | AD0RA2B | adenosine A2b receptor | 0.46 |
| T97105 | TXNRD1 | thioredoxin reductase 1 | 0.52 |
| W79493 | PDE4B | phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) | 0.56 |
| W56577 | VLDLR | very low density lipoprotein receptor | 0.56 |
| AA043909 | PTK2 | PTK2 protein tyrosine kinase 2 | 0.57 |
| T75436 | MAPK10 | mitogen-activated protein kinase 10 | 0.59 |
| N31391 | AKAP12 | A kinase (PRKA) anchor protein (gravin) 12 | 0.62 |
| AA001104 | RGS3 | regulator of G-protein signalling 3 | 0.62 |
| W35223 | ARHGDIB | Rho GDP dissociation inhibitor (GDI) beta | 0.62 |
| H15389 | NPY1R | neuropeptide Y receptor Y1 | 0.63 |
| W52156 | OXTR | oxytocin receptor | 0.63 |
| AA133259 | GEM | GTP binding protein overexpressed in skeletal muscle | 0.64 |
| AA056159 | RHO | rhodopsin (opsin 2, rod pigment) (retinitis pigmentosa 4, autosomal dominant) | 0.64 |
| R52300 | GABRA1 | gamma-aminobutyric acid (GABA) A receptor, alpha 1 | 0.65 |
| BF971558 | STMN1 | stathmin 1/oncoprotein 18 | 1.78 |
| Transcription factors and transcriptional regulators | |||
| W05657 | ELF1 | E74-like factor 1 (ets domain transcription factor) | 0.04 |
| AA044787 | CNOT8 | CCR4-NOT transcription complex, subunit 8 | 0.12 |
| AA127003 | HSZFP36 | ZFP-36 for a zinc finger protein | 0.18 |
| R59543 | NRIP1 | nuclear receptor interacting protein 1 | 0.23 |
| AA005274 | ZNF7 | zinc finger protein 7 (KOX 4, clone HF.16) | 0.24 |
| R35310 | MEIS2 | Meis1, myeloid ecotropic viral integration site 1 homolog 2 (mouse) | 0.24 |
| R20489 | ZNF16 | zinc finger protein 16 (KOX 9) | 0.25 |
| R80355 | BACH1 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 | 0.31 |
| R13988 | EGR3 | early growth response 3 | 0.32 |
| T77424 | FHX | (FOXJ2) forkhead box J2 | 0.38 |
| T84358 | ZNF135 | zinc finger protein 135 (clone pHZ-17) | 0.41 |
| H14414 | ZNF195 | DKFZp434A2314 5′, mRNA sequence. | 0.42 |
| N31546 | IFI16 | interferon, gamma-inducible protein 16 | 0.43 |
| AA043380 | HOXD10 | homeo box D10 | 0.45 |
| N23578 | TCF12 | transcription factor 12 (HTF4, helix-loop-helix transcription factors 4) | 0.49 |
| W90498 | SSB | Sjogren syndrome antigen B (autoantigen La) | 0.52 |
| AA035361 | STAT1 | signal transducer and activator of transcription 1, 91kDa | 0.53 |
| N94714 | YY1 | YY1 transcription factor | 0.54 |
| H50875 | F0XA3 | forkhead box A3 | 0.56 |
| W32908 | FOXO1A | forkhead box O1A (rhabdomyosarcoma) | 0.61 |
| W38673 | ID2 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 0.63 |
| AA043457 | ZNF137 | zinc finger protein 137 (clone pHZ-30) | 0.64 |
| H27140 | ZNF35 | zinc finger protein 35 (clone HF.10) | 0.65 |
| N34004 | STAT6 | signal transducer and activator of transcription 6, interleukin-4 induced | 0.65 |
| R66676 | ETV5 | ets variant gene 5 (ets-related molecule) | 0.65 |
| W16724 | MLL | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | 0.67 |
| Others | |||
| W03376 | NNMT | nicotinamide N-methyltransferase | 0.34 |
| T78107 | GRM5 | glutamate receptor, metabotropic 5 | 0.59 |
| W39552 | BMPR1A | bone morphogenetic protein receptor, type IA | 0.61 |
| BI457624 | PPIL2 | peptidylprolyl isomerase (cyclophilin)-like 2 | 0.62 |
| H92049 | LRBA | LPS-responsive vesicle trafficking, beach and anchor containing | 0.63 |
Table indicates differentially regulated genes from fetal cardiac myocytes treated 12 hours with sera from either healthy or septic individuals. Fold changes are the average ratios between the 4 septic and the 4 healthy individuals.
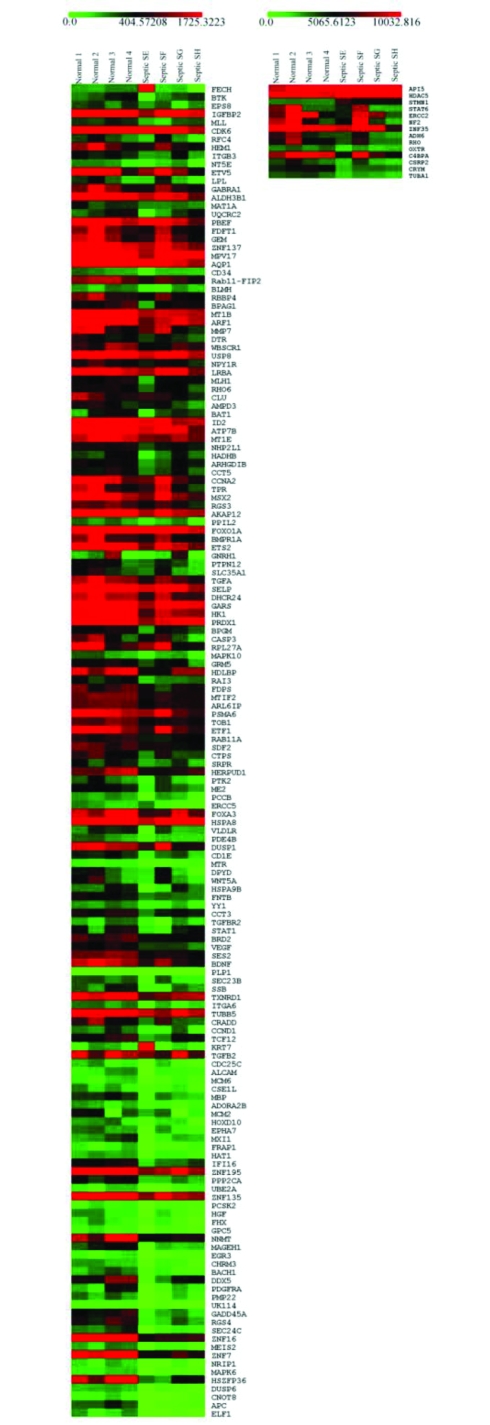
Figure 1.
A summary image showing the ranked differentially expressed genes from fetal cardiac myocytes treated for 12 hours with sera from either healthy or septic individuals. Each column represents the expression level for each sample. Normal indicates myocytes treated with sera from healthy volunteers. Septic indicates cardiac myocytes treated with sera from septic individuals. Gene symbols are indicated on the right hand side. The level of expression is indicated by the colour. High expression levels are coloured red and low expression levels are coloured green as indicated by the scale on the top of the image. This image was created using the multiexperiment viewer of the TM4 microarray software suite.
Cell Cycle Genes
Eight cell cycle related genes were differentially expressed. Seven of these genes were down-regulated ranging from 1.6- to 6.1-fold decrease (including MCM2, MCM6, DUSP1, DUSP6, CCNA2, SESN2, and NHP2L1) and one gene was 1.88-fold up-regulated (HDAC5). Dual specificity protein phosphatases (DUSP) are involved in cellular responses to environmental stress as well as negative regulation of cell proliferation. DUSPs play a role in negatively regulating cell cycle and innate immunity. For example, DUSPs are known to dephosphorylate members of the MAP kinase superfamily and two major kinases which are involved in mediating the inflammatory signaling pathways, the p38 MAP kinase [31] and the JNK [32].
Multiple studies have addressed the critical role of DUSP1 (also known as MAPK phosphatase-1 [MKP-1]) in mediating innate immune responses and cytokine production. [32-35] DUSP1 deficiency was associated with significant increase in mice mortality in LPS-induced septic shock [32, 33]. Chi et al found that DUSP1 deficiency resulted in the sustained activation of p38 MAPK and JNK and was also associated with IL-10 hyperproduction in mice with endotoxic shock [32]. Atrial natriuretic peptide (ANP)-induced DUSP1 activation inhibits cardiac re-programming and the re-expression of fetal protein genes by direct inactivation of P38MAPK [36, 37]. Cyclin A2 (CCNA2) plays an important role in cardiomyocyte mitosis [38] and repair [39]. Expression of CCNA2 is normally silenced in 14 days after birth, suggesting its major role in controlling cardiomyocyte cell cycle and is associated with the decline in cardiac myocyte mitosis [40]. Targeted expression of CCNA2 improved cardiac hemodynamics in myocardial infarction (MI) mouse model [41]. Moreover, CCNA2 transgenic mice showed a remarkable ability to induce cardiac mitosis and repair after MI [39]. Re-expression of DUSP1 and CCNA2 represents a promising therapeutic target that may contribute to improved heart function during sepsis and requires further investigation.
Histones play a principal role in transcriptional regulation, cell cycle progression and developmental events. As a member of the class II histone deacetylase family, HDAC5 possesses histone deacetylase activity and represses transcription. YY1 is a highly conserved transcription factor and is a key regulator of the repressive activity of HDAC5 [42]. YY1 interaction with HDAC5 functions to protect cardiac myocytes against hypertrophy [42]. This interaction prevents HDAC5 cytoplasmic translocation [42]. While located in the nucleus, histone deacetylase 5 interacts with the transcription factor myocyte enhancer factor 2 (MEF2). MEF2 regulates the transcription of the skeletal muscle and cardiac structural genes [43, 44]. HDAC5 interaction with MEF2 results in the inhibition of MEF2 transcriptional activity and the repression of MEF2-dependent genes [45]. Altered Ca2+ signaling, with increased sarcoplasmic reticulum leakage accompanied by a reduction in sarcoplasmic reticulum Ca2+ content is evident during sepsis [46, 47]. Ca2+/calmodulin-dependent protein kinase in the presence of increased levels of cytosolic Ca2+ phosphorylates HDAC5 allowing its translocation to the cytosol [48, 49]. In addition, Ca2+/calmodulin compete for the HDAC5 repressor core inhibiting its binding to MEF2 [45]. The down-regulation of YY1 (1.84-fold down-regulated) results in the cytoplasmic translocation of HDAC5 and the activation of fetal gene programs, like MEF2 [42]. YY1 down-regulation and HDAC5 up-regulation result in HDAC5 nuclear efflux. HDAC5 translocation to the cytosol leads to cardiac hypertrophy through enhanced expression of MEF2-dependent genes.
Mitochondrial Genes
Clear in vivo and in vitro evidence strongly support that sepsis and septic shock severely impair mitochondrial function and cellular oxygen utilization [12]. It was reported that TNF-α induces mitochondrial DNA damage and causes mitochondrial dysfunction in cardiac myocytes via an increase in intracellular oxidative stress [13]. Accordingly, the inhibition of inducible nitric oxide synthase and inducible mitochondrial nitric oxide synthase activities through the administration of melatonin prevented mitochondrial function impairment, restores ATP production and improves survival in CLP mice [50].
Four mitochondrial genes were differentially regulated, three genes were 1.54- to 1.66-fold down-regulated (UQCRC2, HADHB, HK1) and one gene was 2.39-fold up-regulated (FECH). The up-regulation of ferrochelatase (FECH) to our knowledge is not known to contribute to sepsis but its partial deficiency and down-regulation is linked to erythropoietic protoporphyria (inherited heme biosynthesis disorder) [51].
UQCRC2 is a part of the mitochondrial respiratory chain. HADHB, encodes the beta subunit of the mitochondrial trifunctional protein (TFP) and is essential for mitochondrial beta-oxidation of long chain fatty acids. Mutations in this gene are associated with TFP deficiency. Among its clinical phenotypes is severe neonatal presentation with cardiomyopathy. Mild myopathy is the most common phenotype due to this deficiency [52]. Hexokinases commit glucose to the glycolytic pathway through glucose phosphorylation. One member of this family is HK1 and its deficiency is associated with hemolytic anemia [53]. Its expression prevents mitochondrial mediated cell death via permeability transition pore (PTP) closure as well as the accompanying cytochrome c release [54].
Our data indicate that impaired mitochondrial function was evident as HADHB and UQCRC2 were down-regulated. The down-regulation of HK1 may contribute to the activation of mitochondrial-mediated apoptotic pathway observed during sepsis.
Apoptosis Genes
Apoptosis plays a key role in sepsis. In a transgenic mouse model, it was discovered that apoptotic rates of 23 cardiac myocytes per 105 nuclei are sufficient to induce a lethal, dilated cardiomyopathy [55]. In addition, apoptosis leads to a profound immune-paralysis, accompanied by high morbidity and mortality in septic patients as reviewed by Hotchkiss [10].
We observed a 2.2-fold up-regulation of the AP15 gene. Its role in suppressing E2F-dependent apoptosis has been identified and its depletion is lethal to tumor cells [56]. Moreover, cardiac myocytes showed down-regulation of nine genes involved in the positive regulation of apoptosis. One of these is VEGF which is an important cytokine that contributes to sepsis mortality. Increased levels of VEGF has been reported in patients with severe sepsis however lower levels (although higher than healthy controls) were associated with organ dysfunction and poor survival outcome [57]. Two members of the cysteine-aspartic acid protease (caspase) family were also among these nine down-regulated genes (CASP2, CASP3).
Genes related to signaling transduction and transcription
Key signaling molecules such as kinases and transcription factors are modulated after the onset of sepsis. For example, activated p38 MAP kinase/NF-κB signaling molecules play a pivotal role in the sepsis induced conversion of the cardiac myocytes to a proinflammatory phenotype [58]. The pro-inflammatory phenotype is characterized by increased myocyte production of CXC chemokines, specifically keratinocyte-derived chemokine (KC) and LPS-induced chemokine (LIX) and the subsequent transendothelial migration of the polymorphonuclear neutrophils (PMN) [58]. In addition, activation of the JNK pathway in a human in vitro model of sepsis-induced cardiac myocyte apoptosis was evident [11]. Septic sera induced the activation of transcription factors STAT1, IRF1, and NF-κB in human cardiac myocytes [11]. This activation was associated with sepsis induced cellular dysfunction [11].
In our current study, seventeen signaling genes were found to be down-regulated with fold change ranging from 1.55- to 4.96-fold below the normal gene expression in the control samples. One signaling gene, stathmin 1 (STMN1) was 1.78-fold up-regulated, while 26 transcription related genes were down-regulated.
The transcription factor Elf1, a member of Ets family of transcription factors was the most down-regulated gene in our study with 26.49-fold decrease. It was reported that Elf1 plays an important role in regulating transcription and gene expression in lymphoid tissues [59]. Elf1 is also expressed in normal and malignant mammary tissues [60]. Later studies found that endogenous expression of Elf1 and its phosphorylation vary depending on cell type and condition, with higher levels in the hematopoietic cells [61]. In a breast carcinoma study, Elf1 was significantly down-regulated in mouse tumors. Another cancer study reported its role along with p65 in the induction of NFKB1 gene expression and subsequent NFκB activation in specific chemoresistant human T leukemic cell line [62]. Elf1 role in hematopoiesis and transcription regulation in lymphoid cells was previously reviewed [63] but its role in cardiac myocytes and sepsis has yet to be determined.
Among the other down-regulated transcription factors in our study, two belong to the signal transducers and activators of transcription family, STAT1 and STAT6. It is evident that STAT1 plays a pivotal role in cell sensitivity to stress stimuli and stress-induced apoptosis [64]. STAT6 has been shown to have anti-apoptotic activities [65]. CNOT8, also known as POP2, is a transcription factor. It has been previously shown that CNOT8 negatively regulates NF-κB signaling in TNF-α stimulated HEK-293 cell line as it alters the nuclear distribution of p65 [66]. In addition, CNOT8 interferes in the inflammasome-mediated procaspase 1 activation and subsequently obstructs IL-1β secretion [67]. CNOT8 may play a role in protecting the cells from the drastic effects of IL-1β overproduction. CNOT8 is 8.6-fold down-regulated in our study and this may partially account for the increased production and activity of IL-1β in septic patients.
Stathmin 1 (STMN1) plays a pivotal role in mitosis and cell cycle regulation through regulating microtubule dynamics [68]. Dysregulation of stathmin expression has been linked to several pathological conditions. Over-expression has been documented in muliple types of cancer [69-71] and other neuronal disorders [72]. Down-regulation of stathmin has been reported in Alzheimer's disease and Down syndrome [73]. Low levels of STMN1 expression is essential for the proper function of neuronal cells [72, 74], Yamashita et al. conducted a study that addressed the role of activated MAP kinases and the subsequently phosphorylated STMN1 in oxidative stress-induced neuronal cell death [75]. Vancompernolle et al. suggested a role of STMN1 as a downstream target molecule in TNF-induced cell death signaling pathway [76]. We observed a 1.78-fold up-regulation of the STMN1 gene. Increased expression of the STMN1 gene may provide a mechanism for the autonomic dysfunction observed during sepsis as it triggers apoptosis via activated MAP kinases and TNF signaling pathways.
In addition, RGS3, RGS4 and TXNRD1 are important signaling genes, contributing to cardiac hypertrophy and the progression of heart failure by their reduced expression. Recently, the role of RGS4 and RGS3 in the inhibition of ERK1/2 phosphorylation has been confirmed [77]. RGS4 expression is essential in guanylyl cyclase-A (GC-A) - mediated inhibition of cardiac hypertrophy [78]. Heart cells show high levels of VLDL-R expression and its role in lipid metabolism is evident [79]. VLDL-R down-regulation has a crucial role in alterations of heart lipid metabolism observed during sepsis. This alteration is a result of IL-1β signaling and its downstream signaling molecule, Hsp90 [80]. Moreover, enhanced levels of TXNRD1 improve cardiomyocyte survival via its antioxidative activity [81, 82].
Metabolism related genes
Impaired metabolic processes have been observed in septic hearts. Increased expression of iNOS and TNF-α in septic hearts has been linked to distinct structural changes, including lipid accumulation and the disruption of the actin/myosin contractile apparatus [83]. Oxidative damage as well as the altered glucose and lipid metabolism account for these structural changes. Our results confirmed these altered metabolic processes through the down-regulation of 22 metabolism related genes and select members of this group are responsible for lipid and carbohydrate metabolism. We observed the down-regulation of the following lipid metabolism genes; VLDL-R, FDPS, HDLBP, CLU, FDFT1, ALDH3B1, LPL, PCCB. The altered free fatty acid metabolism and uptake by the myocardium in septic patients [18] has been linked to IL-1β mediated down-regulation of genes involved in lipid metabolism [19].
Increased concentrations of free intracellular Ca2+ ions observed during sepsis [46, 47] partially contribute to these metabolic disorders. Sepsis-induced LPL repression contributes to sepsis-associated lipid metabolic disorders in TNF-α-induced Ca2+-dependent manner [84]. The involvement of other lipid metabolism related genes in this study may be of critical importance and requires further investigations. We observed the down-regulation of genes which are involved in carbohydrate metabolism which include; ME2, SLC35A1, and BPGM however the roles that these genes play during sepsis has not been characterized.
Other Genes
Cardiovascular homeostasis is dramatically affected during sepsis. A previous study reported the role that iNOS plays in inducing apoptosis in cardiovascular autonomic centers [16]. As we have previously discussed, activated MAP kinases phosphorylate STMN1 in oxidative stress conditions and induces neuronal cell death [75]. We observed three nervous system related genes that were down-regulated; PLP1, MBP and EPHA7. These three genes have been implicated in nervous system development and maintenance. Two previous studies determined the differential gene expression profile in blood cells derived from children with septic shock and confirmed the role of altered zinc homeostasis in mortality [85, 86]. Consistent with these studies, our results showed a repression in three metal binding genes (MT1E, MT1B, ATP7B).
Our data indicate that septic sera induced cell cycle, metabolic, transcription factor and apoptotic gene expression changes in human myocytes. Mediators in septic sera influence changes in gene expression that lead to organ dysfunction and potentially death. An important mediator is TNF-α, which showed a consistent elevated level in septic sera. In addition, gene expression changes in LPL, STMN1 and mitochondrial genes are shown to be TNF-α-induced. Sepsis-induced differential gene expression of LPL, STMN1 and mitochondrial genes contribute to lipid metabolic disorders, autonomic dysfunction and impaired oxygen utilization, respectively. Moreover, these genes are involved in the activation of mitochondria-mediated apoptotic pathway and subsequently cardiac dysfunction. Members of DUSP and HDAC families, play a key role in protecting cardiac cells from re-programming and the re-expression of fetal gene programs. Targeting DUSP family members, YY1 transcription factor along with HDAC5 may represent a therapeutic approach to improve cardiac function in septic patients. IL-1β, another mediator in septic sera, showed consistent higher levels in septic sera than normal sera. IL-1β was associated with altered lipid metabolism in cardiac myocytes. The role of CNOT8 and Elf1 in NF-κB regulation should be investigated. Furthermore, CNOT8 down-regulation, has shown to contribute to IL-1β increased production in septic patients.
Taken together, our data provide new insights in relation to the changes in gene expression that contribute to cardiac depression and dysfunction observed in septic patients. Identification and characterization of gene expression changes that occur during sepsis may lead to the development of novel therapeutics and diagnostics.
Acknowledgments
The first two authors contributed equally to this paper.
References
- 1.Parrillo JE, Parker MM, Natanson C, Suffredini AF, Danner RL, Cunnion RE, Ognibene FP. Septic shock in humans, advances in the understanding of pathogenesis, cardiovascular dysfunction, and therapy. Ann Intern Med. 1990;113:227–42. doi: 10.7326/0003-4819-113-3-227. [DOI] [PubMed] [Google Scholar]
- 2.Parrillo JE. Pathogenetic mechanisms of septic shock. N Engl J Med. 1993;328:1471–7. doi: 10.1056/NEJM199305203282008. [DOI] [PubMed] [Google Scholar]
- 3.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis, the ACCP/SCCM consensus conference committee, american college of chest Physicians/Society of critical care medicine. Chest. 1992;101:1644–55. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 4.Vincent JL. Cardiovascular alterations in septic shock. J Antimicrob Chemother. 1998;41(Suppl A):9–15. doi: 10.1093/jac/41.suppl_1.9. [DOI] [PubMed] [Google Scholar]
- 5.Zanotti S, Kumar A, Kumar A. Cytokine modulation in sepsis and septic shock. Expert Opin Investig Drugs. 2002;11:1061–75. doi: 10.1517/13543784.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 6.Kumar A, Thota V, Dee L, Olson J, Uretz E, Parrillo JE. Tumor necrosis factor alpha and interleukin 1beta are responsible for in vitro myocardial cell depression induced by human septic shock serum. J Exp Med. 1996;183:949–58. doi: 10.1084/jem.183.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhanantwari P, Nadaraj S, Kenessey A, Chowdhury D, Al-Abed Y, Miller EJ, Ojamaa K. Macrophage migration inhibitory factor induces cardiomyocyte apoptosis. Biochem Biophys Res Commun. 2008;371:298–303. doi: 10.1016/j.bbrc.2008.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flierl MA, Rittirsch D, Huber-Lang MS, Sarma JV, Ward PA. Molecular events in the cardiomyopathy of sepsis. Mol Med. 2008;14:327–36. doi: 10.2119/2007-00130.Flierl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buerke U, Carter JM, Schlitt A, Russ M, Schmidt H, Sibelius U, Grandel U, Grimminger F, Seeger W, Mueller-Werdan U, Werdan K, Buerke M. Apoptosis contributes to septic cardiomyopathy and is improved by simvastatin therapy. Shock. 2008;29:497–503. doi: 10.1097/shk.0b013e318142c434. [DOI] [PubMed] [Google Scholar]
- 10.Hotchkiss RS, Nicholson DW. Apoptosis and caspases regulate death and inflammation in sepsis. Nat Rev Immunol. 2006;6:813–22. doi: 10.1038/nri1943. [DOI] [PubMed] [Google Scholar]
- 11.Kumar A, Kumar A, Michael P, Brabant D, Parissenti AM, Ramana CV, Xu X, Parrillo JE. Human serum from patients with septic shock activates transcription factors STAT1, IRF1, and NF-kappaB and induces apoptosis in human cardiac myocytes. J Biol Chem. 2005;280:42619–26. doi: 10.1074/jbc.M508416200. [DOI] [PubMed] [Google Scholar]
- 12.Crouser ED. Mitochondrial dysfunction in septic shock and multiple organ dysfunction syndrome. Mitochondrion. 2004;4:729–41. doi: 10.1016/j.mito.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 13.Suematsu N, Tsutsui H, Wen J, Kang D, Ikeuchi M, Ide T, Hayashidani S, Shiomi T, Kubota T, Hamasaki N, Takeshita A. Oxidative stress mediates tumor necrosis factor-alpha-induced mitochondrial DNA damage and dysfunction in cardiac myocytes. Circulation. 2003;107:1418–23. doi: 10.1161/01.cir.0000055318.09997.1f. [DOI] [PubMed] [Google Scholar]
- 14.Tavernier B, Garrigue D, Boulle C, Vallet B, Adnet P. Myofilament calcium sensitivity is decreased in skinned cardiac fibres of endotoxin-treated rabbits. Cardiovasc Res. 1998;38:472–9. doi: 10.1016/s0008-6363(98)00028-5. [DOI] [PubMed] [Google Scholar]
- 15.Behrends M, Peters J. The calcium sensitizer levosimendan attenuates endotoxin-evoked myocardial dysfunction in isolated guinea pig hearts. Intensive Care Med. 2003;29:1802–7. doi: 10.1007/s00134-003-1879-8. [DOI] [PubMed] [Google Scholar]
- 16.Sharshar T, Gray F, Lorin de la Grandmaison G, Hopkinson NS, Ross E, Dorandeu A, Orlikowski D, Raphael JC, Gajdos P, Annane D. Apoptosis of neurons in cardiovascular autonomic centres triggered by inducible nitric oxide synthase after death from septic shock. Lancet. 2003;362:1799–805. doi: 10.1016/s0140-6736(03)14899-4. [DOI] [PubMed] [Google Scholar]
- 17.Tessier JP, Thurner B, Jungling E, Luckhoff A, Fischer Y. Impairment of glucose metabolism in hearts from rats treated with endotoxin. Cardiovasc Res. 2003;60:119–30. doi: 10.1016/s0008-6363(03)00320-1. [DOI] [PubMed] [Google Scholar]
- 18.Dhainaut JF, Huyghebaert MF, Monsallier JF, Lefevre G, Dall'Ava-Santucci J, Brunet F, Villemant D, Carli A, Raichvarg D. Coronary hemodynamics and myocardial metabolism of lactate, free fatty acids, glucose, and ketones in patients with septic shock. Circulation. 1987;75:533–41. doi: 10.1161/01.cir.75.3.533. [DOI] [PubMed] [Google Scholar]
- 19.Jia L, Takahashi M, Morimoto H, Takahashi S, Izawa A, Ise H, Iwasaki T, Hattori H, Wu KJ, Ikeda U. Changes in cardiac lipid metabolism during sepsis: The essential role of very low-density lipoprotein receptors. Cardiovasc Res. 2006;69:545–55. doi: 10.1016/j.cardiores.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Okamoto I, Abe M, Shibata K, Shimizu N, Sakata N, Katsuragi T, Tanaka K. Evaluating the role of inducible nitric oxide synthase using a novel and selective inducible nitric oxide synthase inhibitor in septic lung injury produced by cecal ligation and puncture. Am J Respir Crit Care Med. 2000;162:716–22. doi: 10.1164/ajrccm.162.2.9907039. [DOI] [PubMed] [Google Scholar]
- 21.Johnson SB, Lissauer M, Bochicchio GV, Moore R, Cross AS, Scalea TM. Gene expression profiles differentiate between sterile SIRS and early sepsis. Ann Surg. 2007;245:611–21. doi: 10.1097/01.sla.0000251619.10648.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC. The use of gene-expression profiling to identify candidate genes in human sepsis. Am J Respir Crit Care Med. 2007;176:676–84. doi: 10.1164/rccm.200612-1819OC. [DOI] [PubMed] [Google Scholar]
- 23.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis, the ACCP/SCCM consensus conference committee, american college of chest Physicians/Society of critical care medicine. Chest. 1992;101:1644–55. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 24.Villeneuve DJ, Hembruff SL, Veitch Z, Cecchetto M, Dew WA, Parissenti AM. cDNA microarray analysis of isogenic paclitaxel- and doxorubicin-resistant breast tumor cell lines reveals distinct drug-specific genetic signatures of resistance. Breast Cancer Res Treat. 2006;96:17–39. doi: 10.1007/s10549-005-9026-6. [DOI] [PubMed] [Google Scholar]
- 25.Sprowl JA, Villeneuve DJ, Guo B, Young AJ, Hembruff SL, Parissenti AM. Changes in expression of cell wall turnover genes accompany inhibition of chromosome segregation by bovine protein kinase C alpha expression in saccharomyces cerevisiae. Cell Biol Int. 2007;31:1160–72. doi: 10.1016/j.cellbi.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 26.Villeneuve DJ, Parissenti AM. The use of DNA microarrays to investigate the pharmacogenomics of drug response in living systems. Curr Top Med Chem. 2004;4:1329–45. doi: 10.2174/1568026043387610. [DOI] [PubMed] [Google Scholar]
- 27.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: A free, open-source system for microarray data management and analysis. Bio Techniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 28.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 29.Kumar A, Thota V, Dee L, Olson J, Uretz E, Parrillo JE. Tumor necrosis factor alpha and interleukin lbeta are responsible for in vitro myocardial cell depression induced by human septic shock serum. J Exp Med. 1996;183:949–58. doi: 10.1084/jem.183.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parrillo JE, Burch C, Shelhamer JH, Parker MM, Natanson C, Schuette W. A circulating myocardial depressant substance in humans with septic shock, septic shock patients with a reduced ejection fraction have a circulating factor that depresses in vitro myocardial cell performance. J Clin Invest. 1985;76:1539–53. doi: 10.1172/JCI112135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun H, Charles CH, Lau LF, Tonks NK. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell. 1993;75:487–93. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 32.Chi H, Barry SP, Roth RJ, Wu JJ, Jones EA, Bennett AM, Flavell RA. Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proc Natl Acad Sci U S A. 2006;103:2274–9. doi: 10.1073/pnas.0510965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammer M, Mages J, Dietrich H, Servatius A, Howells N, Cato AC, Lang R. Dual specificity phosphatase 1 (DUSP1) regulates a subset of LPS-induced genes and protects mice from lethal endotoxin shock. J Exp Med. 2006;203:15–20. doi: 10.1084/jem.20051753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Q, Wang X, Nelin LD, Yao Y, Matta R, Manson ME, Baliga RS, Meng X, Smith CV, Bauer JA, Chang CH, Liu Y. MAP kinase phosphatase 1 controls innate immune responses and suppresses endotoxic shock. J Exp Med. 2006;203:131–40. doi: 10.1084/jem.20051794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maier JV, Brema S, Tuckermann J, Herzer U, Klein M, Stassen M, Moorthy A, Cato AC. Dual specificity phosphatase 1 knockout mice show enhanced susceptibility to anaphylaxis but are sensitive to glucocorticoids. Mol Endocrinol. 2007;21:2663–71. doi: 10.1210/me.2007-0067. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi D, Kudoh S, Shiojima I, Zou Y, Harada K, Shimoyama M, Imai Y, Monzen K, Yamazaki T, Yazaki Y, Nagai R, Komuro I. Atrial natriuretic peptide inhibits cardiomyocyte hypertrophy through mitogen-activated protein kinase phosphatase-1. Biochem Biophys Res Commun. 2004;322:310–9. doi: 10.1016/j.bbrc.2004.07.119. [DOI] [PubMed] [Google Scholar]
- 37.Fuller SJ, Davies EL, Gillespie-Brown J, Sun H, Tonks NK. Mitogen-activated protein kinase phosphatase 1 inhibits the stimulation of gene expression by hypertrophic agonists in cardiac myocytes. Biochem J. 1997;323(Pt 2):313–9. doi: 10.1042/bj3230313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhry HW, Dashoush NH, Tang H, Zhang L, Wang X, Wu EX, Wolgemuth DJ. Cyclin A2 mediates cardiomyocyte mitosis in the postmitotic myocardium. J Biol Chem. 2004;279:35858–66. doi: 10.1074/jbc.M404975200. [DOI] [PubMed] [Google Scholar]
- 39.Cheng RK, Asai T, Tang H, Dashoush NH, Kara RJ, Costa KD, Naka Y, Wu EX, Wolgemuth DJ, Chaudhry HW. Cyclin A2 induces cardiac regeneration after myocardial infarction and prevents heart failure. Circ Res. 2007;100:1741–8. doi: 10.1161/CIRCRESAHA.107.153544. [DOI] [PubMed] [Google Scholar]
- 40.Yoshizumi M, Lee WS, Hsieh CM, Tsai JC, Li J, Perrella MA, Patterson C, Endege WO, Schlegel R, Lee ME. Disappearance of cyclin A correlates with permanent withdrawal of cardiomyocytes from the cell cycle in human and rat hearts. J Clin Invest. 1995;95:2275–80. doi: 10.1172/JCI117918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo YJ, Panlilio CM, Cheng RK, Liao GP, Suarez EE, Atluri P, Chaudhry HW. Myocardial regeneration therapy for ischemic cardiomyopathy with cyclin A2. J Thorac Cardiovasc Surg. 2007;133:927–33. doi: 10.1016/j.jtcvs.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 42.Sucharov CC, Dockstader K, McKinsey TA. YY1 protects cardiac myocytes from pathologic hypertrophy by interacting with HDAC5. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-12-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–96. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 44.Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci U S A. 1996;93:9366–73. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berger I, Bieniossek C, Schaffitzel C, Hassler M, Santelli E, Richmond TJ. Direct interaction of Ca2+/calmodulin inhibits histone deacetylase 5 repressor core binding to myocyte enhancer factor 2. J Biol Chem. 2003;278:17625–35. doi: 10.1074/jbc.M301646200. [DOI] [PubMed] [Google Scholar]
- 46.Zhu X, Bernecker OY, Manohar NS, Hajjar RJ, Hellman J, Ichinose F, Valdivia HH, Schmidt U. Increased leakage of sarcoplasmic reticulum Ca2+ contributes to abnormal myocyte Ca2+ handling and shortening in sepsis. Crit Care Med. 2005;33:598–604. doi: 10.1097/01.ccm.0000152223.27176.a6. [DOI] [PubMed] [Google Scholar]
- 47.Sayeed MM. Signaling mechanisms of altered cellular responses in trauma, burn, and sepsis: Role of Ca2+ Arch Surg. 2000;135:1432–42. doi: 10.1001/archsurg.135.12.1432. [DOI] [PubMed] [Google Scholar]
- 48.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–40. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passier R, Zeng H, Frey N, Naya FJ, Nicol RL, McKinsey TA, Overbeek P, Richardson JA, Grant SR, Olson EN. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J Clin Invest. 2000;105:1395–406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Escames G, Lopez LC, Ortiz F, Lopez A, Garcia JA, Ros E, Acuna-Castroviejo D. Attenuation of cardiac mitochondrial dysfunction by melatonin in septic mice. FEBS J. 2007;274:2135–47. doi: 10.1111/j.1742-4658.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- 51.Gouya L, Martin-Schmitt C, Robreau AM, Austerlitz F, Da Silva V, Brun P, Simonin S, Lyoumi S, Grandchamp B, Beaumont C, Puy H, Deybach JC. Contribution of a common single-nucleotide polymorphism to the genetic predisposition for erythropoietic protoporphyria. Am J Hum Genet. 2006;78:2–14. doi: 10.1086/498620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spiekerkoetter U, Sun B, Khuchua Z, Bennett MJ, Strauss AW. Molecular and phenotypic heterogeneity in mitochondrial trifunctional protein deficiency due to beta-subunit mutations. Hum Mutat. 2003;21:598–607. doi: 10.1002/humu.10211. [DOI] [PubMed] [Google Scholar]
- 53.Peters LL, Lane PW, Andersen SG, Gwynn B, Barker JE, Beutler E. Downeast anemia (dea), a new mouse model of severe nonspherocytic hemolytic anemia caused by hexokinase (HK(1)) deficiency. Blood Cells Mol Dis. 2001;27:850–60. doi: 10.1006/bcmd.2001.0454. [DOI] [PubMed] [Google Scholar]
- 54.Azoulay-Zohar H, Israelson A, Abu-Hamad S, Shoshan-Barmatz V. In self-defence: Hexokinase promotes voltage-dependent anion channel closure and prevents mitochondria-mediated apoptotic cell death. Biochem J. 2004;377:347–55. doi: 10.1042/BJ20031465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wencker D, Chandra M, Nguyen K, Miao W, Garantziotis S, Factor SM, Shirani J, Armstrong RC, Kitsis RN. A mechanistic role for cardiac myocyte apoptosis in heart failure. J Clin Invest. 2003;111:1497–504. doi: 10.1172/JCI17664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morris EJ, Michaud WA, Ji JY, Moon NS, Rocco JW, Dyson NJ. Functional identification of Api5 as a suppressor of E2F-dependent apoptosis in vivo. PLoS Genet. 2006;2:el96. doi: 10.1371/journal.pgen.0020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karlsson S, Pettila V, Tenhunen J, Lund V, Hovilehto S, Ruokonen E, Finnsepsis Study Group Vascular endothelial growth factor in severe sepsis and septic shock. Anesth Analg. 2008;106:1820–6. doi: 10.1213/ane.0b013e31816a643f. [DOI] [PubMed] [Google Scholar]
- 58.Yang M, Wu J, Martin CM, Kvietys PR, Rui T. Important role of p38 MAP kinase/NF-kappaB signaling pathway in the sepsis-induced conversion of cardiac myocytes to a proinflammatory phenotype. Am J Physiol Heart Circ Physiol. 2008;294:H994–1001. doi: 10.1152/ajpheart.01044.2007. [DOI] [PubMed] [Google Scholar]
- 59.Thompson CB, Wang CY, Ho IC, Bohjanen PR, Petryniak B, June CH, Miesfeldt S, Zhang L, Nabel GJ, Karpinski B. Cis-acting sequences required for inducible interleukin-2 enhancer function bind a novel ets-related protein, elf-1. Mol Cell Biol. 1992;12:1043–53. doi: 10.1128/mcb.12.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galang CK, Muller WJ, Foos G, Oshima RG, Hauser CA. Changes in the expression of many ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004;279:11281–92. doi: 10.1074/jbc.M311887200. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi K, Hayashi N, Shimokawa T, Umehara N, Kaminogawa S, Ra C. Cooperative regulation of fc receptor gamma-chain gene expression by multiple transcription factors, including Sp1, GABP, and elf-1. J Biol Chem. 2008;283:15134–41. doi: 10.1074/jbc.M800498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang PY, Miyamoto S. Nuclear factor-kappaB dimer exchange promotes a p21(waf1/cip1) superinduction response in human T leukemic cells. Mol Cancer Res. 2006;4:101–12. doi: 10.1158/1541-7786.MCR-05-0259. [DOI] [PubMed] [Google Scholar]
- 63.Gallant S, Gilkeson G. ETS transcription factors and regulation of immunity. Arch Immunol Ther Exp (Warsz) 2006;54:149–63. doi: 10.1007/s00005-006-0017-z. [DOI] [PubMed] [Google Scholar]
- 64.Janjua S, Stephanou A, Latchman DS. The C-terminal activation domain of the STAT-1 transcription factor is necessary and sufficient for stress-induced apoptosis. Cell Death Differ. 2002;9:1140–6. doi: 10.1038/sj.cdd.4401082. [DOI] [PubMed] [Google Scholar]
- 65.Zhang WJ, Li BH, Yang XZ, Li PD, Yuan Q, Liu XH, Xu SB, Zhang Y, Yuan J, Gerhard GS, Masker KK, Dong C, Koltun WA, Chorney MJ. IL-4-induced Stat6 activities affect apoptosis and gene expression in breast cancer cells. Cytokine. 2008;42:39–47. doi: 10.1016/j.cyto.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 66.Bedoya F, Sandler LL, Harton JA. Pyrin-only protein 2 modulates NF-kappaB and disrupts ASC:CLR interactions. J Immunol. 2007;178:3837–45. doi: 10.4049/jimmunol.178.6.3837. [DOI] [PubMed] [Google Scholar]
- 67.Dorfleutner A, Bryan NB, Talbott SJ, Funya KN, Rellick SL, Reed JC, Shi X, Rojanasakul Y, Flynn DC, Stehlik C. Cellular pyrin domain-only protein 2 is a candidate regulator of inflammasome activation. Infect Immun. 2007;75:1484–92. doi: 10.1128/IAI.01315-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–31. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- 69.Chang CL, Hora N, Huberman N, Hinderer R, Kukuruga M, Hanash SM. Oncoprotein 18 levels and phosphorylation mediate megakaryocyte polyploidization in human erythroleukemia cells. Proteomics. 2001;1:1415–23. doi: 10.1002/1615-9861(200111)1:11<1415::AID-PROT1415>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 70.Singer S, Ehemann V, Brauckhoff A, Keith M, Vreden S, Schirmacher P, Breuhahn K. Protumorigenic overexpression of stathmin/Op18 by gain-of-function mutation in p53 in human hepatocarcinogenesis. Hepatology. 2007;46:759–68. doi: 10.1002/hep.21736. [DOI] [PubMed] [Google Scholar]
- 71.Wei SH, Lin F, Wang X, Gao P, Zhang HZ. Prognostic significance of stathmin expression in correlation with metastasis and clinicopathological characteristics in human ovarian carcinoma. Acta Histochem. 2008;110:59–65. doi: 10.1016/j.acthis.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 72.Liu A, Stadelmann C, Moscarello M, Bruck W, Sobel A, Mastronardi FG, Casaccia-Bonnefil P. Expression of stathmin, a developmentally controlled cytoskeleton-regulating molecule, in demyelinating disorders. J Neurosci. 2005;25:737–47. doi: 10.1523/JNEUROSCI.4174-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheon MS, Fountoulakis M, Cairns NJ, Dierssen M, Herkner K, Lubec G. Decreased protein levels of stathmin in adult brains with down syndrome and alzheimer's disease. J Neural Transm Suppl. 2001;(61):281–8. doi: 10.1007/978-3-7091-6262-0_23. [DOI] [PubMed] [Google Scholar]
- 74.Ohkawa N, Fujitani K, Tokunaga E, Furuya S, Inokuchi K. The microtubule destabilizer stathmin mediates the development of dendritic arbors in neuronal cells. J Cell Sci. 2007;120:1447–56. doi: 10.1242/jcs.001461. [DOI] [PubMed] [Google Scholar]
- 75.Yamashita H, Nakamura K, Arai H, Furumoto H, Fujimoto M, Kashiwagi S, Morimatsu M. Electrophoretic studies on the phosphorylation of stathmin and mitogen-activated protein kinases in neuronal cell death induced by oxidized very-low-density lipoprotein with apolipoprotein E. Electrophoresis. 2002;23:998–1004. doi: 10.1002/1522-2683(200204)23:7/8<998::AID-ELPS998>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 76.Vancompernolle K, Boonefaes T, Mann M, Fiers W, Grooten J. Tumor necrosis factor-induced microtubule stabilization mediated by hyperphosphorylated oncoprotein 18 promotes cell death. J Biol Chem. 2000;275:33876–82. doi: 10.1074/jbc.M004785200. [DOI] [PubMed] [Google Scholar]
- 77.Anger T, Klintworth N, Stumpf C, Daniel WG, Mende U, Garlichs CD. RGS protein specificity towards gq- and Gi/o-mediated ERK 1/2 and akt activation, in vitro. J Biochem Mol Biol. 2007;40:899–910. doi: 10.5483/bmbrep.2007.40.6.899. [DOI] [PubMed] [Google Scholar]
- 78.Tokudome T, Kishimoto I, Horio T, Arai Y, Schwenke DO, Hino J, Okano I, Kawano Y, Kohno M, Miyazato M, Nakao K, Kangawa K. Regulator of G-protein signaling subtype 4 mediates antihypertrophic effect of locally secreted natriuretic peptides in the heart. Circulation. 2008;117:2329–39. doi: 10.1161/CIRCULATIONAHA.107.732990. [DOI] [PubMed] [Google Scholar]
- 79.Takahashi S, Kawarabayasi Y, Nakai T, Sakai J, Yamamoto T. Rabbit very low density lipoprotein receptor: A low density lipoprotein receptor-like protein with distinct ligand specificity. Proc Natl Acad Sci U S A. 1992;89:9252–6. doi: 10.1073/pnas.89.19.9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jia L, Takahashi M, Morimoto H, Takahashi S, Izawa A, Ise H, Iwasaki T, Hattori H, Wu KJ, Ikeda U. Changes in cardiac lipid metabolism during sepsis: The essential role of very low-density lipoprotein receptors. Cardiovasc Res. 2006;69:545–55. doi: 10.1016/j.cardiores.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 81.Giraud MN, Fluck M, Zuppinger C, Suter TM. Expressional reprogramming of survival pathways in rat cardiocytes by neuregulin-1beta. J Appl Physiol. 2005;99:313–22. doi: 10.1152/japplphysiol.00609.2004. [DOI] [PubMed] [Google Scholar]
- 82.Satoh M, Matter CM, Ogita H, Takeshita K, Wang CY, Dorn GW, 2nd, Liao JK. Inhibition of apoptosis-regulated signaling kinase-1 and prevention of congestive heart failure by estrogen. Circulation. 2007;115:3197–204. doi: 10.1161/CIRCULATIONAHA.106.657981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rossi MA, Celes MR, Prado CM, Saggioro FP. Myocardial structural changes in long-term human severe sepsis/septic shock may be responsible for cardiac dysfunction. Shock. 2007;27:10–8. doi: 10.1097/01.shk.0000235141.05528.47. [DOI] [PubMed] [Google Scholar]
- 84.Sakaguchi S, Iizuka Y, Furusawa S, Takayanagi M, Satoh S. Preventive effects of a verapamil against tumor necrosis factor-alpha-induced shock symptoms: Approached from lipoprotein metabolic disorders. Int Immunopharmacol. 2002;2:867–73. doi: 10.1016/s1567-5769(02)00009-7. [DOI] [PubMed] [Google Scholar]
- 85.Cvijanovich N, Shanley TP, Lin R, Allen GL, Thomas NJ, Checchia P, Anas N, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Wong HR, Genomics of Pediatric SIRS/Septic Shock Investigators Validating the genomic signature of pediatric septic shock. Physiol Genomics. 2008;34:127–34. doi: 10.1152/physiolgenomics.00025.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Tagavilla MA, Odoms K, Dunsmore K, Barnes M, Aronow BJ, Genomics of Pediatric SIRS/Septic Shock Investigators Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–55. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]