Table 3.
Average (±SD) values of the kinetic parameters together with their range given between brackets for the reversible two-tissue compartment model
| Data | K1 | k2 | k3 | 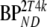 |
VT | 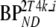 |
Vb |
|---|---|---|---|---|---|---|---|
| HC ctx | 0.25 ± 0.03 (0.2–0.3) | 0.06 ± 0.01 (0.04–0.08) | 0.02 ± 0.01 (0.01–0.05) | 2.14 ± 0.71 (1–4) | 14 ± 6 (7–25) | 0.22 ± 0.18 (−0.04–0.7) | 0.03 ± 0.01 (0.01–0.05) |
| AD ctx | 0.36 ± 0.08 (0.3–0.5) | 0.06 ± 0.01 (0.04–0.08) | 0.02 ± 0.01 (0.01–0.04) | 1.76 ± 0.71 (1–3) | 17 ± 5 (9–26) | 0.23 ± 0.20 (−0.03–0.6) | 0.05 ± 0.03 (0.02–0.11) |
| AD + HC cer | 0.35 ± 0.09 (0.3–0.5) | 0.07 ± 0.01 (0.06–0.09) | 0.02 ± 0.02 (0.01–0.06) | 1.67 ± 0.56 (1–3) | 13 ± 5 (6–20) | – | 0.03 ± 0.02 (0.01–0.07) |
These consisted of several typical AD cortical (ctx, i.e., orbital frontal cortex, medial inferior frontal cortex, anterior cingulate cortex, superior temporal cortex, parietal cortex, medial inferior temporal cortex, superior frontal cortex, enthorinal cortex, hippocampus) and cerebellar (cer) gray matter regions from healthy controls (HC) and AD subjects. K1 and K1m are given in ml·cm−3·min−1, k2 and k3 in min−1. 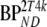 and
and 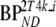 are binding potentials estimated directly and indirectly, respectively, using the reversible two-tissue plasma input model.
are binding potentials estimated directly and indirectly, respectively, using the reversible two-tissue plasma input model. 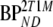 and
and 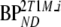 are binding potentials estimated directly and indirectly, respectively, using the metabolite model.
are binding potentials estimated directly and indirectly, respectively, using the metabolite model. 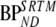 is the binding potential estimated using the simplified reference tissue model
is the binding potential estimated using the simplified reference tissue model
